Reduced pathogenicity of the SARS-CoV-2 omicron variant in hamsters
- PMID: 35313451
- PMCID: PMC8926874
- DOI: 10.1016/j.medj.2022.03.004
Reduced pathogenicity of the SARS-CoV-2 omicron variant in hamsters
Abstract
Background: The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron (B.1.1.529) variant has proven to be highly transmissible and has outcompeted the Delta variant in many regions of the world. Early reports have also suggested that Omicron may result in less severe clinical disease in humans. Here, we show that Omicron is less pathogenic than prior SARS-CoV-2 variants in Syrian golden hamsters.
Methods: Hamsters were inoculated with either SARS-CoV-2 Omicron or other SARS-CoV-2 variants. Animals were followed for weight loss, and upper and lower respiratory tract tissues were assessed for viral loads and histopathology.
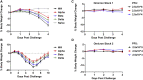
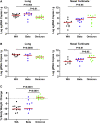
Findings: Infection of hamsters with the SARS-CoV-2 WA1/2020, Alpha, Beta, or Delta strains led to 4%-10% weight loss by day 4 and 10%-17% weight loss by day 6. In contrast, infection of hamsters with two different Omicron challenge stocks did not result in any detectable weight loss, even at high challenge doses. Omicron infection led to substantial viral replication in both the upper and lower respiratory tracts but demonstrated lower viral loads in lung parenchyma and reduced pulmonary pathology compared with WA1/2020 infection.
Conclusions: These data suggest that the SARS-CoV-2 Omicron variant may result in robust upper respiratory tract infection, but less severe lower respiratory tract clinical disease, compared with prior SARS-CoV-2 variants.
Funding: Funding for this study was provided by NIH grant CA260476, the Massachusetts Consortium for Pathogen Readiness, the Ragon Institute, and the Musk Foundation.
Keywords: COVID-19; SARS-CoV-2; hamster; omicron; pathogenicity.
© 2022 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L., et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in golden syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. - DOI - PMC - PubMed
-
- Tostanoski L.H., Yu J., Mercado N.B., McMahan K., Jacob-Dolan C., Martinot A.J., Piedra-Mora C., Anioke T., Chang A., Giffin V.M., et al. Immunity elicited by natural infection or Ad26.COV2.S vaccination protects hamsters against SARS-CoV-2 variants of concern. Sci. Transl. Med. 2021;13:eabj3789. doi: 10.1126/scitranslmed.abj3789. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

