This is a preprint.
Biomarkers Selection for Population Normalization in SARS-CoV-2 Wastewater-based Epidemiology
- PMID: 35313587
- PMCID: PMC8936110
- DOI: 10.1101/2022.03.14.22272359
Biomarkers Selection for Population Normalization in SARS-CoV-2 Wastewater-based Epidemiology
Update in
-
Biomarkers selection for population normalization in SARS-CoV-2 wastewater-based epidemiology.Water Res. 2022 Sep 1;223:118985. doi: 10.1016/j.watres.2022.118985. Epub 2022 Aug 15. Water Res. 2022. PMID: 36030667 Free PMC article.
Abstract
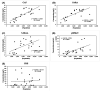
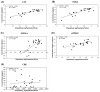
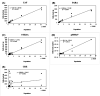
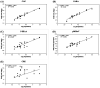
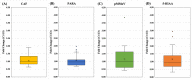
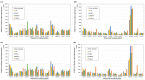
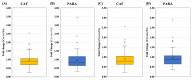
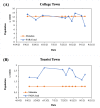
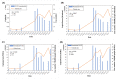
Wastewater-based epidemiology (WBE) has been one of the most cost-effective approaches to track the SARS-CoV-2 levels in the communities since the COVID-19 outbreak in 2020. Normalizing SARS-CoV-2 concentrations by the population biomarkers in wastewater can be critical for interpreting the viral loads, comparing the epidemiological trends among the sewersheds, and identifying the vulnerable communities. In this study, five population biomarkers, pepper mild mottle virus (pMMoV), creatinine (CRE), 5-hydroxyindoleacetic acid (5-HIAA), caffeine (CAF) and its metabolite paraxanthine (PARA) were investigated for their utility in normalizing the SARS-CoV-2 loads through developed direct and indirect approaches. Their utility in assessing the real-time population contributing to the wastewater was also evaluated. The best performed candidate was further tested for its capacity for improving correlation between normalized SARS-CoV-2 loads and the clinical cases reported in the City of Columbia, Missouri, a university town with a constantly fluctuated population. Our results showed that, except CRE, the direct and indirect normalization approaches using biomarkers allow accounting for the changes in wastewater dilution and differences in relative human waste input over time regardless flow volume and population at any given WWTP. Among selected biomarkers, PARA is the most reliable population biomarker in determining the SARS-CoV-2 load per capita due to its high accuracy, low variability, and high temporal consistency to reflect the change in population dynamics and dilution in wastewater. It also demonstrated its excellent utility for real-time assessment of the population contributing to the wastewater. In addition, the viral loads normalized by the PARA-estimated population significantly improved the correlation ( rho =0.5878, p <0.05) between SARS-CoV-2 load per capita and case numbers per capita. This chemical biomarker offers an excellent alternative to the currently CDC-recommended pMMoV genetic biomarker to help us understand the size, distribution, and dynamics of local populations for forecasting the prevalence of SARS-CoV2 within each sewershed.
Highlight bullet points: The paraxanthine (PARA), the metabolite of the caffeine, is a more reliable population biomarker in SARS-CoV-2 wastewater-based epidemiology studies than the currently recommended pMMoV genetic marker.SARS-CoV-2 load per capita could be directly normalized using the regression functions derived from correlation between paraxanthine and population without flowrate and population data.Normalizing SARS-CoV-2 levels with the chemical marker PARA significantly improved the correlation between viral loads per capita and case numbers per capita.The chemical marker PARA demonstrated its excellent utility for real-time assessment of the population contributing to the wastewater.
Figures


References
-
- WHO Naming the coronavirus disease (COVID-19) and the virus that causes it. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technica....
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
