This is a preprint.
Parsing the role of NSP1 in SARS-CoV-2 infection
- PMID: 35313595
- PMCID: PMC8936099
- DOI: 10.1101/2022.03.14.484208
Parsing the role of NSP1 in SARS-CoV-2 infection
Update in
-
Parsing the role of NSP1 in SARS-CoV-2 infection.Cell Rep. 2022 Jun 14;39(11):110954. doi: 10.1016/j.celrep.2022.110954. Epub 2022 May 26. Cell Rep. 2022. PMID: 35671758 Free PMC article.
Abstract
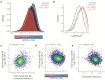
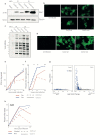
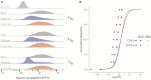
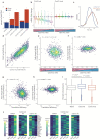
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the cause of the ongoing coronavirus disease 19 (COVID-19) pandemic. Despite its urgency, we still do not fully understand the molecular basis of SARS-CoV-2 pathogenesis and its ability to antagonize innate immune responses. SARS-CoV-2 leads to shutoff of cellular protein synthesis and over-expression of nsp1, a central shutoff factor in coronaviruses, inhibits cellular gene translation. However, the diverse molecular mechanisms nsp1 employs as well as its functional importance in infection are still unresolved. By overexpressing various nsp1 mutants and generating a SARS-CoV-2 mutant in which nsp1 does not bind ribosomes, we untangle the effects of nsp1. We uncover that nsp1, through inhibition of translation and induction of mRNA degradation, is the main driver of host shutoff during SARS-CoV-2 infection. Furthermore, we find the propagation of nsp1 mutant virus is inhibited specifically in cells with intact interferon (IFN) response as well as in-vivo , in infected hamsters, and this attenuation is associated with stronger induction of type I IFN response. This illustrates that nsp1 shutoff activity has an essential role mainly in counteracting the IFN response. Overall, our results reveal the multifaceted approach nsp1 uses to shut off cellular protein synthesis and uncover the central role it plays in SARS-CoV-2 pathogenesis, explicitly through blockage of the IFN response.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
