Fusarivirus accessory helicases present an evolutionary link for viruses infecting plants and fungi
- PMID: 35314402
- PMCID: PMC9243621
- DOI: 10.1016/j.virs.2022.03.010
Fusarivirus accessory helicases present an evolutionary link for viruses infecting plants and fungi
Abstract
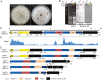
A significant number of mycoviruses have been identified that are related to plant viruses, but their evolutionary relationships are largely unexplored. A fusarivirus, Rhizoctonia solani fusarivirus 4 (RsFV4), was identified in phytopathogenic fungus Rhizoctonia solani (R. solani) strain XY74 co-infected by an alphaendornavirus. RsFV4 had a genome of 10,833 nt (excluding the poly-A tail), and consisted of four non-overlapping open reading frames (ORFs). ORF1 encodes an 825 aa protein containing a conserved helicase domain (Hel1). ORF3 encodes 1550 aa protein with two conserved domains, namely an RNA-dependent RNA polymerase (RdRp) and another helicase (Hel2). The ORF2 and ORF4 likely encode two hypothetical proteins (520 and 542 aa) with unknown functions. The phylogenetic analysis based on Hel2 and RdRp suggest that RsFV4 was positioned within the fusarivirus group, but formed an independent branch with three previously reported fusariviruses of R. solani. Notably, the Hel1 and its relatives were phylogenetically closer to helicases of potyviruses and hypoviruses than fusariviruses, suggesting fusarivirus Hel1 formed an evolutionary link between these three virus groups. This finding provides evidence of the occurrence of a horizontal gene transfer or recombination event between mycoviruses and plant viruses or between mycoviruses. Our findings are likely to enhance the understanding of virus evolution and diversity.
Keywords: Fusarivirus; Helicase; Horizontal gene transfer (HGT); Hypovirus; Potyvirus.
Copyright © 2022 The Authors. Publishing services by Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Complete genome sequence of a novel fusarivirus from Rhizoctonia solani AG-3 PT strain 3P-2-2.Arch Virol. 2023 Jan 27;168(2):75. doi: 10.1007/s00705-022-05673-7. Arch Virol. 2023. PMID: 36707487
-
Molecular characterization of a novel fusarivirus infecting the plant-pathogenic fungus Alternaria solani.Arch Virol. 2021 Jul;166(7):2063-2067. doi: 10.1007/s00705-021-05105-y. Epub 2021 May 13. Arch Virol. 2021. PMID: 33983501
-
Molecular characterization of a novel fusarivirus infecting the edible fungus Auricularia heimuer.Arch Virol. 2020 Nov;165(11):2689-2693. doi: 10.1007/s00705-020-04781-6. Epub 2020 Aug 18. Arch Virol. 2020. PMID: 32812093
-
Mycoviruses as a part of the global virome: Diversity, evolutionary links and lifestyle.Adv Virus Res. 2023;115:1-86. doi: 10.1016/bs.aivir.2023.02.002. Epub 2023 Apr 21. Adv Virus Res. 2023. PMID: 37173063 Review.
-
Mycoviruses in Fusarium Species: An Update.Front Cell Infect Microbiol. 2019 Jul 18;9:257. doi: 10.3389/fcimb.2019.00257. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31380300 Free PMC article. Review.
Cited by
-
A group of segmented viruses contains genome segments sharing homology with multiple viral taxa.J Virol. 2025 Jul 22;99(7):e0033225. doi: 10.1128/jvi.00332-25. Epub 2025 Jun 4. J Virol. 2025. PMID: 40464578 Free PMC article.
-
ICTV Virus Taxonomy Profile: Fusariviridae 2024.J Gen Virol. 2024 Apr;105(4):001973. doi: 10.1099/jgv.0.001973. J Gen Virol. 2024. PMID: 38619867 Free PMC article.
References
-
- Ballut L., Marchadier B., Baguet A., Tomasetto C., Séraphin B., Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005;12:861–869. - PubMed
-
- Bonfante P., Genre A. Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat. Commun. 2010;1:1–11. - PubMed
-
- Bräutigam A., Mullick T., Schliesky S., Weber A.P. Critical assessment of assembly strategies for non-model species mRNA-Seq data and application of next-generation sequencing to the comparison of C3 and C4 species. J. Exp. Bot. 2011;62:3093–3102. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

