Rational design of hairpin RNA excited states reveals multi-step transitions
- PMID: 35314698
- PMCID: PMC8938425
- DOI: 10.1038/s41467-022-29194-8
Rational design of hairpin RNA excited states reveals multi-step transitions
Abstract
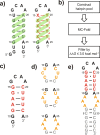
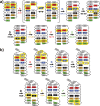
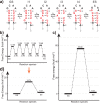
RNA excited states represent a class of high-energy-level and thus low-populated conformational states of RNAs that are sequestered within the free energy landscape until being activated by cellular cues. In recent years, there has been growing interest in structural and functional studies of these transient states, but the rational design of excited states remains unexplored. Here we developed a method to design small hairpin RNAs with predefined excited states that exchange with ground states through base pair reshuffling, and verified these transient states by combining NMR relaxation dispersion technique and imino chemical shift prediction. Using van't Hoff analysis and accelerated molecular dynamics simulations, a mechanism of multi-step sequential transition has been revealed. The efforts made in this study will expand the scope of RNA rational design, and also contribute towards improved predictions of RNA secondary structure.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Characterizing RNA Excited States Using NMR Relaxation Dispersion.Methods Enzymol. 2015;558:39-73. doi: 10.1016/bs.mie.2015.02.002. Epub 2015 Mar 25. Methods Enzymol. 2015. PMID: 26068737 Free PMC article.
-
Probing invisible, low-populated States of protein molecules by relaxation dispersion NMR spectroscopy: an application to protein folding.Acc Chem Res. 2008 Mar;41(3):442-51. doi: 10.1021/ar700189y. Epub 2008 Feb 15. Acc Chem Res. 2008. PMID: 18275162 Review.
-
Studying sparsely populated conformational states in RNA combining chemical synthesis and solution NMR spectroscopy.Methods. 2018 Sep 15;148:39-47. doi: 10.1016/j.ymeth.2018.05.007. Epub 2018 May 31. Methods. 2018. PMID: 29753787
-
NMR Relaxation Dispersion Methods for the Structural and Dynamic Analysis of Quickly Interconverting, Low-Populated Conformational Substates.Methods Mol Biol. 2022;2376:187-203. doi: 10.1007/978-1-0716-1716-8_11. Methods Mol Biol. 2022. PMID: 34845611
-
Mutate-and-chemical-shift-fingerprint (MCSF) to characterize excited states in RNA using NMR spectroscopy.Nat Protoc. 2021 Nov;16(11):5146-5170. doi: 10.1038/s41596-021-00606-1. Epub 2021 Oct 4. Nat Protoc. 2021. PMID: 34608336 Review.
Cited by
-
4D Assembly of Time-dependent Lanthanide Supramolecular Multicolor Phosphorescence for Encryption and Visual Sensing.Adv Sci (Weinh). 2025 Apr;12(14):e2415418. doi: 10.1002/advs.202415418. Epub 2025 Feb 14. Adv Sci (Weinh). 2025. PMID: 39950854 Free PMC article.
-
Visualizing a two-state conformational ensemble in stem-loop 3 of the transcriptional regulator 7SK RNA.Nucleic Acids Res. 2024 Jan 25;52(2):940-952. doi: 10.1093/nar/gkad1159. Nucleic Acids Res. 2024. PMID: 38084902 Free PMC article.
-
Stick-slip unfolding favors self-association of expanded HTT mRNA.bioRxiv [Preprint]. 2024 Jun 3:2024.05.31.596809. doi: 10.1101/2024.05.31.596809. bioRxiv. 2024. Update in: Nat Commun. 2024 Oct 9;15(1):8738. doi: 10.1038/s41467-024-52764-x. PMID: 38895475 Free PMC article. Updated. Preprint.
-
Stick-slip unfolding favors self-association of expanded HTT mRNA.Nat Commun. 2024 Oct 9;15(1):8738. doi: 10.1038/s41467-024-52764-x. Nat Commun. 2024. PMID: 39384800 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

