Development of spirulina for the manufacture and oral delivery of protein therapeutics
- PMID: 35314813
- PMCID: PMC9200632
- DOI: 10.1038/s41587-022-01249-7
Development of spirulina for the manufacture and oral delivery of protein therapeutics
Erratum in
-
Author Correction: Development of spirulina for the manufacture and oral delivery of protein therapeutics.Nat Biotechnol. 2022 Jun;40(6):974. doi: 10.1038/s41587-022-01323-0. Nat Biotechnol. 2022. PMID: 35440781 Free PMC article. No abstract available.
Abstract
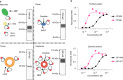
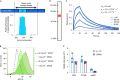
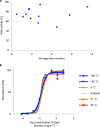
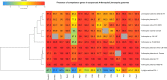
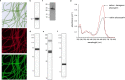
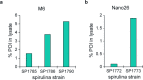
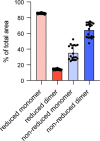
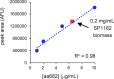
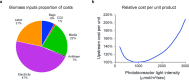
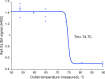
The use of the edible photosynthetic cyanobacterium Arthrospira platensis (spirulina) as a biomanufacturing platform has been limited by a lack of genetic tools. Here we report genetic engineering methods for stable, high-level expression of bioactive proteins in spirulina, including large-scale, indoor cultivation and downstream processing methods. Following targeted integration of exogenous genes into the spirulina chromosome (chr), encoded protein biopharmaceuticals can represent as much as 15% of total biomass, require no purification before oral delivery and are stable without refrigeration and protected during gastric transit when encapsulated within dry spirulina. Oral delivery of a spirulina-expressed antibody targeting campylobacter-a major cause of infant mortality in the developing world-prevents disease in mice, and a phase 1 clinical trial demonstrated safety for human administration. Spirulina provides an advantageous system for the manufacture of orally delivered therapeutic proteins by combining the safety of a food-based production host with the accessible genetic manipulation and high productivity of microbial platforms.
© 2022. The Author(s).
Conflict of interest statement
J.R. and B.F. are the founders and current employees of Lumen Bioscience, Inc. (Lumen) and own stock/stock options in Lumen. B.W.J., H.Z., M.G., T.A., D.T.B., J.A., N.K., R.K., C.G., J.F., T.P., C. Brady, S.E., M.Z., A.P., J.L., M.T., T.S., D.D., J.M., L.G., J.D., D.F., A.M., B.K., K.S. and C. Behnke are current employees or paid advisors of Lumen; all current and former employees own stock/stock options of Lumen. K.C., R.L., D.C., N.S., E.A., T.N., A.R., A.T., A.K. and B.R. were employees of Lumen at the time of data generation. Lumen has issued patents (US no. 10,131,870) and a pending patent application (International Application no. PCT/US2020/040794) relating to certain research described in this article.
Figures




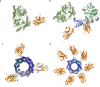

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

