Enzymatic assembly of the salinosporamide γ-lactam-β-lactone anticancer warhead
- PMID: 35314816
- PMCID: PMC9058210
- DOI: 10.1038/s41589-022-00993-w
Enzymatic assembly of the salinosporamide γ-lactam-β-lactone anticancer warhead
Abstract
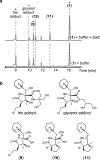
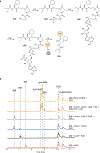
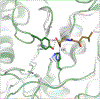
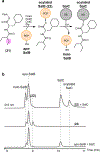
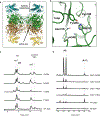
The marine microbial natural product salinosporamide A (marizomib) is a potent proteasome inhibitor currently in clinical trials for the treatment of brain cancer. Salinosporamide A is characterized by a complex and densely functionalized γ-lactam-β-lactone bicyclic warhead, the assembly of which has long remained a biosynthetic mystery. Here, we report an enzymatic route to the salinosporamide core catalyzed by a standalone ketosynthase (KS), SalC. Chemoenzymatic synthesis of carrier protein-tethered substrates, as well as intact proteomics, allowed us to probe the reactivity of SalC and understand its role as an intramolecular aldolase/β-lactone synthase with roles in both transacylation and bond-forming reactions. Additionally, we present the 2.85-Å SalC crystal structure that, combined with site-directed mutagenesis, allowed us to propose a bicyclization reaction mechanism. This work challenges our current understanding of the role of KS enzymes and establishes a basis for future efforts toward streamlined production of a clinically relevant chemotherapeutic.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
COMPETING INTERESTS
The authors declare no competing interests.
Figures
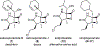










Similar articles
-
Salinosporamide natural products: Potent 20 S proteasome inhibitors as promising cancer chemotherapeutics.Angew Chem Int Ed Engl. 2010 Dec 3;49(49):9346-67. doi: 10.1002/anie.201000728. Angew Chem Int Ed Engl. 2010. PMID: 20927786 Free PMC article. Review.
-
Bacterial self-resistance to the natural proteasome inhibitor salinosporamide A.ACS Chem Biol. 2011 Nov 18;6(11):1257-64. doi: 10.1021/cb2002544. Epub 2011 Sep 26. ACS Chem Biol. 2011. PMID: 21882868 Free PMC article.
-
A mechanistic and kinetic study of the beta-lactone hydrolysis of Salinosporamide A (NPI-0052), a novel proteasome inhibitor.J Pharm Sci. 2007 Aug;96(8):2037-47. doi: 10.1002/jps.20835. J Pharm Sci. 2007. PMID: 17554770
-
Generating a generation of proteasome inhibitors: from microbial fermentation to total synthesis of salinosporamide a (marizomib) and other salinosporamides.Mar Drugs. 2010 Mar 25;8(4):835-80. doi: 10.3390/md8040835. Mar Drugs. 2010. PMID: 20479958 Free PMC article. Review.
-
Structural Insights into Salinosporamide a Mediated Inhibition of the Human 20S Proteasome.Molecules. 2025 Mar 20;30(6):1386. doi: 10.3390/molecules30061386. Molecules. 2025. PMID: 40142161 Free PMC article.
Cited by
-
Alligamycin A, an antifungal β-lactone spiroketal macrolide from Streptomyces iranensis.Nat Commun. 2024 Oct 26;15(1):9259. doi: 10.1038/s41467-024-53695-3. Nat Commun. 2024. PMID: 39461983 Free PMC article.
-
Ketosynthase Domain Catalyzes β-Lactonization in the Biosynthesis of the HMG-CoA Synthase Inhibitor Hymeglusin.J Am Chem Soc. 2025 Jul 23;147(29):25136-25141. doi: 10.1021/jacs.5c07060. Epub 2025 Jul 10. J Am Chem Soc. 2025. PMID: 40635350 Free PMC article.
-
Biosynthesis of lactacystin as a proteasome inhibitor.Commun Chem. 2025 Jan 13;8(1):9. doi: 10.1038/s42004-025-01406-4. Commun Chem. 2025. PMID: 39806036 Free PMC article.
-
Identification of γ-butyrolactone signalling molecules in diverse actinomycetes using resin-assisted isolation and chemoenzymatic synthesis.RSC Chem Biol. 2025 Feb 25;6(4):630-641. doi: 10.1039/d5cb00007f. eCollection 2025 Apr 2. RSC Chem Biol. 2025. PMID: 40046449 Free PMC article.
-
Antibacterial Polyketides Isolated from the Marine-Derived Fungus Fusarium solani 8388.J Fungi (Basel). 2023 Aug 24;9(9):875. doi: 10.3390/jof9090875. J Fungi (Basel). 2023. PMID: 37754983 Free PMC article.
References
-
- Roth P et al. A phase III trial of marizomib in combination with temozolomide-based radiochemotherapy versus temozolomide-based radiochemotherapy alone in patients with newly diagnosed glioblastoma. Am. J. Clin. Oncol 39, 2004–2004 (2021).
-
- Feling RH et al. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed 42, 355–357 (2003). - PubMed
-
- Williams PG et al. New cytotoxic salinosporamides from the marine Actinomycete Salinispora tropica. J. Org. Chem 70, 6196–6203 (2005). - PubMed
-
- Macherla VR et al. Structure-activity relationship studies of salinosporamide A (NPI-0052), a novel marine derived proteasome inhibitor. J. Med. Chem 48, 3684–3687 (2005). - PubMed
METHODS REFERENCES
-
- Gilchrist CLM & Chooi Y-H clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics (2021). - PubMed
-
- Kieser T Practical Streptomyces Genetics. (John Innes Foundation, 2000).
-
- MacNeil DJ et al. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111, 61–68 (1992). - PubMed
-
- Flett F, Mersinias V & Smith CP High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting Streptomycetes. FEMS Microbiol. Lett 155, 223–229 (1997). - PubMed

