Whole-genome risk prediction of common diseases in human preimplantation embryos
- PMID: 35314819
- PMCID: PMC8938270
- DOI: 10.1038/s41591-022-01735-0
Whole-genome risk prediction of common diseases in human preimplantation embryos
Abstract
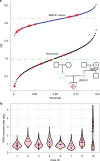
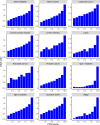
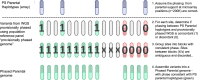
Preimplantation genetic testing (PGT) of in-vitro-fertilized embryos has been proposed as a method to reduce transmission of common disease; however, more comprehensive embryo genetic assessment, combining the effects of common variants and rare variants, remains unavailable. Here, we used a combination of molecular and statistical techniques to reliably infer inherited genome sequence in 110 embryos and model susceptibility across 12 common conditions. We observed a genotype accuracy of 99.0-99.4% at sites relevant to polygenic risk scoring in cases from day-5 embryo biopsies and 97.2-99.1% in cases from day-3 embryo biopsies. Combining rare variants with polygenic risk score (PRS) magnifies predicted differences across sibling embryos. For example, in a couple with a pathogenic BRCA1 variant, we predicted a 15-fold difference in odds ratio (OR) across siblings when combining versus a 4.5-fold or 3-fold difference with BRCA1 or PRS alone. Our findings may inform the discussion of utility and implementation of genome-based PGT in clinical practice.
© 2022. The Author(s).
Conflict of interest statement
A.K., K.I., J.S., O.S., T.T., D.H. and M.R. are either current or previous employees of, and M.B., G.G., P.C.N., B.L. and L.G. are either current or previous consultants with, MyOme. M.R., D.K., M.B. and M.K. are current employees of Natera. The remaining authors declare no competing interests.
Figures




Comment in
-
Polygenic embryo testing: understated ethics, unclear utility.Nat Med. 2022 Mar;28(3):446-448. doi: 10.1038/s41591-022-01743-0. Nat Med. 2022. PMID: 35314818 Free PMC article.
-
The alarming rise of complex genetic testing in human embryo selection.Nature. 2022 Mar;603(7902):549-550. doi: 10.1038/d41586-022-00787-z. Nature. 2022. PMID: 35314828 No abstract available.
Similar articles
-
Screening embryos for polygenic disease risk: a review of epidemiological, clinical, and ethical considerations.Hum Reprod Update. 2024 Oct 1;30(5):529-557. doi: 10.1093/humupd/dmae012. Hum Reprod Update. 2024. PMID: 38805697 Free PMC article. Review.
-
Preimplantation genetic testing for aneuploidies (abnormal number of chromosomes) in in vitro fertilisation.Cochrane Database Syst Rev. 2020 Sep 8;9(9):CD005291. doi: 10.1002/14651858.CD005291.pub3. Cochrane Database Syst Rev. 2020. PMID: 32898291 Free PMC article.
-
Development of an artificial intelligence model for predicting the likelihood of human embryo euploidy based on blastocyst images from multiple imaging systems during IVF.Hum Reprod. 2022 Jul 30;37(8):1746-1759. doi: 10.1093/humrep/deac131. Hum Reprod. 2022. PMID: 35674312 Free PMC article.
-
Genome sequencing of human in vitro fertilisation embryos for pathogenic variation screening.Sci Rep. 2020 Mar 2;10(1):3795. doi: 10.1038/s41598-020-60704-0. Sci Rep. 2020. PMID: 32123222 Free PMC article.
-
A whole-genome sequencing-based novel preimplantation genetic testing method for de novo mutations combined with chromosomal balanced translocations.J Assist Reprod Genet. 2020 Oct;37(10):2525-2533. doi: 10.1007/s10815-020-01921-4. Epub 2020 Aug 11. J Assist Reprod Genet. 2020. PMID: 32783137 Free PMC article.
Cited by
-
The future of nephrology in 2050.Future Healthc J. 2025 Mar 31;12(1):100236. doi: 10.1016/j.fhj.2025.100236. eCollection 2025 Mar. Future Healthc J. 2025. PMID: 40236931 Free PMC article.
-
Inconsistent embryo selection across polygenic score methods.Nat Hum Behav. 2024 Dec;8(12):2264-2267. doi: 10.1038/s41562-024-02019-y. Nat Hum Behav. 2024. PMID: 39402258 No abstract available.
-
Promises and pitfalls of preimplantation genetic testing for polygenic disorders: a narrative review.F S Rev. 2025 Jun;6(1):100085. doi: 10.1016/j.xfnr.2024.100085. Epub 2024 Dec 19. F S Rev. 2025. PMID: 40213363
-
Foretelling the Future: Preimplantation Genetic Testing and the Coming of Polygenic Embryo Screening.J Clin Med. 2025 May 31;14(11):3885. doi: 10.3390/jcm14113885. J Clin Med. 2025. PMID: 40507647 Free PMC article. Review.
-
SHaploseek is a sequencing-only, high-resolution method for comprehensive preimplantation genetic testing.Sci Rep. 2023 Oct 21;13(1):18036. doi: 10.1038/s41598-023-45292-z. Sci Rep. 2023. PMID: 37865712 Free PMC article.
References
-
- Handyside AH, et al. Validation and first clinical application of karyomapping for preimplantation diagnosis (PGD) of Gaucher disease combined with 24 chromosome screening. Fertil. Steril. 2010;94:S79–S80. doi: 10.1016/j.fertnstert.2010.07.309. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

