PSMA-targeting TGFβ-insensitive armored CAR T cells in metastatic castration-resistant prostate cancer: a phase 1 trial
- PMID: 35314843
- PMCID: PMC10308799
- DOI: 10.1038/s41591-022-01726-1
PSMA-targeting TGFβ-insensitive armored CAR T cells in metastatic castration-resistant prostate cancer: a phase 1 trial
Abstract
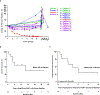
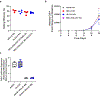
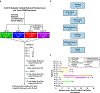
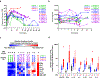
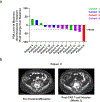
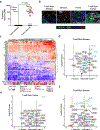
Chimeric antigen receptor (CAR) T cells have demonstrated promising efficacy, particularly in hematologic malignancies. One challenge regarding CAR T cells in solid tumors is the immunosuppressive tumor microenvironment (TME), characterized by high levels of multiple inhibitory factors, including transforming growth factor (TGF)-β. We report results from an in-human phase 1 trial of castration-resistant, prostate cancer-directed CAR T cells armored with a dominant-negative TGF-β receptor (NCT03089203). Primary endpoints were safety and feasibility, while secondary objectives included assessment of CAR T cell distribution, bioactivity and disease response. All prespecified endpoints were met. Eighteen patients enrolled, and 13 subjects received therapy across four dose levels. Five of the 13 patients developed grade ≥2 cytokine release syndrome (CRS), including one patient who experienced a marked clonal CAR T cell expansion, >98% reduction in prostate-specific antigen (PSA) and death following grade 4 CRS with concurrent sepsis. Acute increases in inflammatory cytokines correlated with manageable high-grade CRS events. Three additional patients achieved a PSA reduction of ≥30%, with CAR T cell failure accompanied by upregulation of multiple TME-localized inhibitory molecules following adoptive cell transfer. CAR T cell kinetics revealed expansion in blood and tumor trafficking. Thus, clinical application of TGF-β-resistant CAR T cells is feasible and generally safe. Future studies should use superior multipronged approaches against the TME to improve outcomes.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests
Patents, Royalties, Other Intellectual Property: S.F.L., M.M.D, D.L.S., C.H.J. and J.A.F. have filed patent applications in the field of T-cell therapy for cancer and have received royalties. C.H.J. and A.C. are co-founders of Tmunity Therapeutics. M.M.D. has received research funding from Tmunity Therapeutics and serves on the Scientific Advisory Board for Cellares Corporation. S.F.L. has served as a consultant for Novartis Pharmaceuticals, Kite Pharma, and Wugen and receives clinical trial funding from Novartis Pharmaceuticals. J.A.F. is a member of the Scientific Advisory Boards of Cartography Bio. and Shennon Biotechnologies Inc. The remaining authors declare no competing interests.
Figures
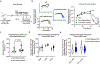




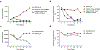
Comment in
-
CAR T cells reach clinical milestone in prostate cancer.Nat Med. 2022 Apr;28(4):635-636. doi: 10.1038/s41591-022-01742-1. Nat Med. 2022. PMID: 35314844 No abstract available.
-
Genetically engineered CAR T cells to hack prostate cancer TME.Nat Rev Urol. 2022 May;19(5):255. doi: 10.1038/s41585-022-00599-w. Nat Rev Urol. 2022. PMID: 35411068 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

