Multiresolution nondestructive 3D pathology of whole lymph nodes for breast cancer staging
- PMID: 35315258
- PMCID: PMC8936940
- DOI: 10.1117/1.JBO.27.3.036501
Multiresolution nondestructive 3D pathology of whole lymph nodes for breast cancer staging
Abstract
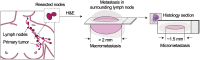
Significance: For breast cancer patients, the extent of regional lymph node (LN) metastasis influences the decision to remove all axillary LNs. Metastases are currently identified and classified with visual analysis of a few thin tissue sections with conventional histology that may underrepresent the extent of metastases.
Aim: We sought to enable nondestructive three-dimensional (3D) pathology of human axillary LNs and to develop a practical workflow for LN staging with our method. We also sought to evaluate whether 3D pathology improves staging accuracy in comparison to two-dimensional (2D) histology.
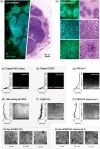
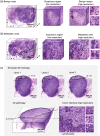
Approach: We developed a method to fluorescently stain and optically clear LN specimens for comprehensive imaging with multiresolution open-top light-sheet microscopy. We present an efficient imaging and data-processing workflow for rapid evaluation of H&E-like datasets in 3D, with low-resolution screening to identify potential metastases followed by high-resolution localized imaging to confirm malignancy.
Results: We simulate LN staging with 3D and 2D pathology datasets from 10 metastatic nodes, showing that 2D pathology consistently underestimates metastasis size, including instances in which 3D pathology would lead to upstaging of the metastasis with important implications on clinical treatment.
Conclusions: Our 3D pathology method may improve clinical management for breast cancer patients by improving staging accuracy of LN metastases.
Keywords: breast cancer; lymph node staging; open-top light-sheet microscopy; three-dimensional pathology.
Figures


References
-
- Hortobagyi G. N., et al. , AJCC Cancer Staging Manual, Vol. 48(8), Springer International Publishing; (2017).
-
- Mamounas E. P., et al. , “Effect of serial sectioning and immunohistochemistry (IHC) on sentinel lymph nodes (SLNs) on the false-negative rate (FNR) of SLN biopsy (SLNB): Results from NSABP B-32,” J. Clin. Oncol. 29(27), 86 (2011).JCONDN10.1200/jco.2011.29.27_suppl.86 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

