Genomic Characteristics of Recently Recognized Vibrio cholerae El Tor Lineages Associated with Cholera in Bangladesh, 1991 to 2017
- PMID: 35315699
- PMCID: PMC9045249
- DOI: 10.1128/spectrum.00391-22
Genomic Characteristics of Recently Recognized Vibrio cholerae El Tor Lineages Associated with Cholera in Bangladesh, 1991 to 2017
Abstract
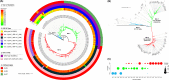
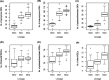
Comparative genomic analysis of Vibrio cholerae El Tor associated with endemic cholera in Asia revealed two distinct lineages, one dominant in Bangladesh and the other in India. An in-depth whole-genome study of V. cholerae El Tor strains isolated during endemic cholera in Bangladesh (1991 to 2017) included reference genome sequence data obtained online. Core genome phylogeny established using single nucleotide polymorphisms (SNPs) showed V. cholerae El Tor strains comprised two lineages, BD-1 and BD-2, which, according to Bayesian phylodynamic analysis, originated from paraphyletic group BD-0 around 1981. BD-1 and BD-2 lineages overlapped temporally but were negatively associated as causative agents of cholera during 2004 to 2017. Genome-wide association study (GWAS) revealed 140 SNPs and 31 indels, resulting in gene alleles unique to BD-1 and BD-2. Regression analysis of root to tip distance and year of isolation indicated early BD-0 strains at the base, whereas BD-1 and BD-2 subsequently emerged and progressed by accumulating SNPs. Pangenome analysis provided evidence of gene acquisition by both BD-1 and BD-2, of which six crucial proteins of known function were predominant in BD-2. BD-1 and BD-2 diverged and have distinctively different genomic traits, namely, heterogeneity in VSP-2, VPI-1, mobile elements, toxin encoding elements, and total gene abundance. In addition, the observed phage-inducible chromosomal island-like element (PLE1), and SXT ICE elements (ICETET) in BD-2 presumably provided a fitness advantage for the lineage to outcompete BD-1 as the etiological agent of endemic cholera in Bangladesh, with implications for global cholera epidemiology. IMPORTANCE Cholera is a global disease with specific reference to the Bay of Bengal Ganges Delta where Vibrio cholerae O1 El Tor, the causative agent of the disease showed two circulating lineages, one dominant in Bangladesh and the other in India. Results of an in-depth genomic study of V. cholerae associated with endemic cholera during the past 27 years (1991 to 2017) indicate emergence and succession of the two lineages, BD-1 and BD-2, arising from a common ancestral paraphyletic group, BD-0, comprising the early strains and short-term evolution of the bacterium in Bangladesh. Among the two V. cholerae lineages, BD-2 supersedes BD-1 and is predominant in the most recent endemic cholera in Bangladesh. The BD-2 lineage contained significantly more SNPs and indels, and showed richness in gene abundance, including antimicrobial resistance genes, gene cassettes, and PLE to fight against bacteriophage infection, acquired over time. These findings have important epidemic implications on a global scale.
Keywords: Vibrio cholerae lineages; antimicrobial resistance; bacterial evolution; comparative genomics; phage-inducible chromosomal island-like elements (PLE).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Vibrio cholerae lineage and pangenome diversity vary geographically across Bangladesh over 1 year.Microb Genom. 2025 Jul;11(7). doi: 10.1099/mgen.0.001437. Microb Genom. 2025. PMID: 40709915
-
Genetic approach toward linkage of Iran 2012-2016 cholera outbreaks with 7th pandemic Vibrio cholerae.BMC Microbiol. 2024 Jan 22;24(1):33. doi: 10.1186/s12866-024-03185-9. BMC Microbiol. 2024. PMID: 38254012 Free PMC article.
-
Vibrio cholerae lineage and pangenome diversity varies geographically across Bangladesh over one year.bioRxiv [Preprint]. 2024 Nov 14:2024.11.12.623281. doi: 10.1101/2024.11.12.623281. bioRxiv. 2024. Update in: Microb Genom. 2025 Jul;11(7). doi: 10.1099/mgen.0.001437. PMID: 39605465 Free PMC article. Updated. Preprint.
-
Global status of antimicrobial resistance among environmental isolates of Vibrio cholerae O1/O139: a systematic review and meta-analysis.Antimicrob Resist Infect Control. 2022 Apr 25;11(1):62. doi: 10.1186/s13756-022-01100-3. Antimicrob Resist Infect Control. 2022. PMID: 35468830 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
Cited by
-
Water pollution, cholera, and the role of probiotics: a comprehensive review in relation to public health in Bangladesh.Front Microbiol. 2025 Jan 14;15:1523397. doi: 10.3389/fmicb.2024.1523397. eCollection 2024. Front Microbiol. 2025. PMID: 39877756 Free PMC article. Review.
-
Prevalent chromosome fusion in Vibrio cholerae O1.Nat Commun. 2025 Jul 1;16(1):5830. doi: 10.1038/s41467-025-60699-0. Nat Commun. 2025. PMID: 40593602 Free PMC article.
-
Gut microbiota shifts favorably with delivery of handwashing with soap and water treatment intervention in a prospective cohort (CHoBI7 trial).J Health Popul Nutr. 2023 Dec 21;42(1):146. doi: 10.1186/s41043-023-00477-0. J Health Popul Nutr. 2023. PMID: 38129922 Free PMC article. Clinical Trial.
-
National Hospital-Based Sentinel Surveillance for Cholera in Bangladesh: Epidemiological Results from 2014 to 2021.Am J Trop Med Hyg. 2023 Aug 14;109(3):575-583. doi: 10.4269/ajtmh.23-0074. Print 2023 Sep 6. Am J Trop Med Hyg. 2023. PMID: 37580033 Free PMC article.
-
A fatal case of Vibrio cholerae-associated diarrhea and bacteremia in a 30-year-old carrier of beta-thalassemia.Gut Pathog. 2024 Dec 19;16(1):76. doi: 10.1186/s13099-024-00655-3. Gut Pathog. 2024. PMID: 39702517 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

