Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases
- PMID: 35316014
- PMCID: PMC7617139
- DOI: 10.1021/acschembio.1c00732
Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases
Abstract
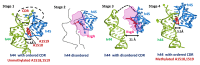
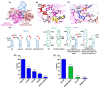
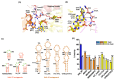
Methylation of specific nucleotides is integral for ribosomal biogenesis and also serves as a common mechanism to confer antibiotic resistance by pathogenic bacteria. Here, by determining the high-resolution structure of the 30S-KsgA complex by cryo-electron microscopy, a state was captured, where KsgA juxtaposes between helices h44 and h45 of the 30S ribosome, separating them, thereby enabling remodeling of the surrounded rRNA and allowing the cognate site to enter the methylation pocket. With the structure as a guide, several mutant versions of the ribosomes, where interacting bases in the catalytic helix h45 and surrounding helices h44, h24, and h27, were mutated and evaluated for their methylation efficiency revealing factors that direct the enzyme to its cognate site with high fidelity. The biochemical studies show that the three-dimensional environment of the ribosome enables the interaction of select loop regions in KsgA with the ribosome helices paramount to maintain selectivity.
Figures





References
-
- Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12:35–48. - PubMed
-
- Arenz S, Wilson DN. Blast from the Past: Reassessing Forgotten Translation Inhibitors, Antibiotic Selectivity, and Resistance Mechanisms to Aid Drug Development. Mol Cell. 2016;61:3–14. - PubMed
-
- Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42–51. - PubMed
-
- Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

