PHGDH is required for germinal center formation and is a therapeutic target in MYC-driven lymphoma
- PMID: 35316216
- PMCID: PMC9057607
- DOI: 10.1172/JCI153436
PHGDH is required for germinal center formation and is a therapeutic target in MYC-driven lymphoma
Abstract
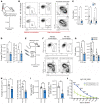
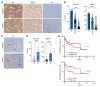
The synthesis of serine from glucose is a key metabolic pathway supporting cellular proliferation in healthy and malignant cells. Despite this, the role that this aspect of metabolism plays in germinal center biology and pathology is not known. Here, we performed a comprehensive characterization of the role of the serine synthesis pathway in germinal center B cells and lymphomas derived from these cells. We demonstrate that upregulation of a functional serine synthesis pathway is a metabolic hallmark of B cell activation and the germinal center reaction. Inhibition of phosphoglycerate dehydrogenase (PHGDH), the first and rate-limiting enzyme in this pathway, led to defective germinal formation and impaired high-affinity antibody production. In addition, overexpression of enzymes involved in serine synthesis was a characteristic of germinal center B cell-derived lymphomas, with high levels of expression being predictive of reduced overall survival in diffuse large B cell lymphoma. Inhibition of PHGDH induced apoptosis in lymphoma cells, reducing disease progression. These findings establish PHGDH as a critical player in humoral immunity and a clinically relevant target in lymphoma.
Keywords: Amino acid metabolism; Immunoglobulins; Immunology; Lymphomas; Metabolism.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

