Rapamycin limits CD4+ T cell proliferation in simian immunodeficiency virus-infected rhesus macaques on antiretroviral therapy
- PMID: 35316218
- PMCID: PMC9106346
- DOI: 10.1172/JCI156063
Rapamycin limits CD4+ T cell proliferation in simian immunodeficiency virus-infected rhesus macaques on antiretroviral therapy
Abstract
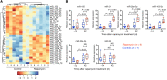
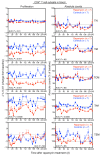
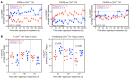
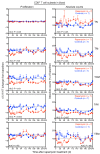
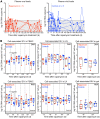
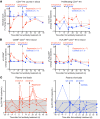
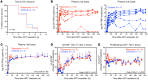
Proliferation of latently infected CD4+ T cells with replication-competent proviruses is an important mechanism contributing to HIV persistence during antiretroviral therapy (ART). One approach to targeting this latent cell expansion is to inhibit mTOR, a regulatory kinase involved with cell growth, metabolism, and proliferation. Here, we determined the effects of chronic mTOR inhibition with rapamycin with or without T cell activation in SIV-infected rhesus macaques (RMs) on ART. Rapamycin perturbed the expression of multiple genes and signaling pathways important for cellular proliferation and substantially decreased the frequency of proliferating CD4+ memory T cells (TM cells) in blood and tissues. However, levels of cell-associated SIV DNA and SIV RNA were not markedly different between rapamycin-treated RMs and controls during ART. T cell activation with an anti-CD3LALA antibody induced increases in SIV RNA in plasma of RMs on rapamycin, consistent with SIV production. However, upon ART cessation, both rapamycin and CD3LALA-treated and control-treated RMs rebounded in less than 12 days, with no difference in the time to viral rebound or post-ART viral load set points. These results indicate that, while rapamycin can decrease the proliferation of CD4+ TM cells, chronic mTOR inhibition alone or in combination with T cell activation was not sufficient to disrupt the stability of the SIV reservoir.
Keywords: AIDS/HIV; Homeostasis; T cells.
Conflict of interest statement
Figures


Similar articles
-
Antibody-Mediated CD4 Depletion Induces Homeostatic CD4+ T Cell Proliferation without Detectable Virus Reactivation in Antiretroviral Therapy-Treated Simian Immunodeficiency Virus-Infected Macaques.J Virol. 2018 Oct 29;92(22):e01235-18. doi: 10.1128/JVI.01235-18. Print 2018 Nov 15. J Virol. 2018. PMID: 30185596 Free PMC article.
-
Bone Marrow-Derived CD4+ T Cells Are Depleted in Simian Immunodeficiency Virus-Infected Macaques and Contribute to the Size of the Replication-Competent Reservoir.J Virol. 2018 Dec 10;93(1):e01344-18. doi: 10.1128/JVI.01344-18. Print 2019 Jan 1. J Virol. 2018. PMID: 30305357 Free PMC article.
-
Initiation of Antiretroviral Therapy Restores CD4+ T Memory Stem Cell Homeostasis in Simian Immunodeficiency Virus-Infected Macaques.J Virol. 2016 Jul 11;90(15):6699-6708. doi: 10.1128/JVI.00492-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27170752 Free PMC article.
-
Brain macrophages harbor latent, infectious simian immunodeficiency virus.AIDS. 2019 Dec 1;33 Suppl 2(Suppl 2):S181-S188. doi: 10.1097/QAD.0000000000002269. AIDS. 2019. PMID: 31789817 Free PMC article. Review.
-
HIV Persistence in Adipose Tissue Reservoirs.Curr HIV/AIDS Rep. 2018 Feb;15(1):60-71. doi: 10.1007/s11904-018-0378-z. Curr HIV/AIDS Rep. 2018. PMID: 29423731 Free PMC article. Review.
Cited by
-
Quantitative and Qualitative Distinctions between HIV-1 and SIV Reservoirs: Implications for HIV-1 Cure-Related Studies.Viruses. 2024 Mar 27;16(4):514. doi: 10.3390/v16040514. Viruses. 2024. PMID: 38675857 Free PMC article. Review.
-
Sequence Analysis of microRNAs Encoded by Simian Lymphocryptoviruses.Viruses. 2024 Dec 16;16(12):1923. doi: 10.3390/v16121923. Viruses. 2024. PMID: 39772230 Free PMC article.
-
Rapamycin enhances CAR-T control of HIV replication and reservoir elimination in vivo.J Clin Invest. 2025 Feb 11;135(7):e185489. doi: 10.1172/JCI185489. J Clin Invest. 2025. PMID: 39932788 Free PMC article.
-
Recent advances of the mammalian target of rapamycin signaling in mesenchymal stem cells.Front Genet. 2022 Aug 30;13:970699. doi: 10.3389/fgene.2022.970699. eCollection 2022. Front Genet. 2022. PMID: 36110206 Free PMC article. Review.
-
HIV-1 replication and latency are balanced by mTOR-driven cell metabolism.Front Cell Infect Microbiol. 2022 Nov 17;12:1068436. doi: 10.3389/fcimb.2022.1068436. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36467738 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

