Profilin is involved in G1 to S phase progression and mitotic spindle orientation during Leishmania donovani cell division cycle
- PMID: 35316283
- PMCID: PMC8939790
- DOI: 10.1371/journal.pone.0265692
Profilin is involved in G1 to S phase progression and mitotic spindle orientation during Leishmania donovani cell division cycle
Abstract
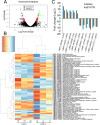
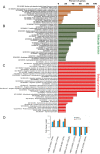
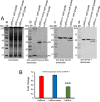
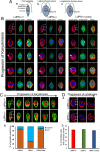
Profilin is a multi-ligand binding protein, which is a key regulator of actin dynamics and involved in regulating several cellular functions. It is present in all eukaryotes, including trypanosomatids such as Leishmania. However, not much is known about its functions in these organisms. Our earlier studies have shown that Leishmania parasites express a single homologue of profilin (LdPfn) that binds actin, phosphoinositides and poly- L- proline motives, and depletion of its intracellular pool to 50%of normal levels affects the cell growth and intracellular trafficking. Here, we show, employing affinity pull-down and mass spectroscopy, that LdPfn interacted with a large number of proteins, including those involved in mRNA processing and protein translation initiation, such as eIF4A1. Further, we reveal, using mRNA Seq analysis, that depletion of LdPfn in Leishmania cells (LdPfn+/-) resulted in significantly reduced expression of genes which encode proteins involved in cell cycle regulation, mRNA translation initiation, nucleosides and amino acids transport. In addition, we show that in LdPfn+/- cells, cellular levels of eIF4A1 protein were significantly decreased, and during their cell division cycle, G1-to-S phase progression was delayed and orientation of mitotic spindle altered. These changes were, however, reversed to normal by episomal expression of GFP-LdPfn in LdPfn+/- cells. Taken together, our results indicate that profilin is involved in regulation of G1-to-S phase progression and mitotic spindle orientation in Leishmania cell cycle, perhaps through its interaction with elF4A1 protein.
Conflict of interest statement
The authors of this manuscript have no conflicts of interest to declare.
Figures


Similar articles
-
Actin sequestering protein, profilin, regulates intracellular vesicle transport in Leishmania.Mol Biochem Parasitol. 2020 Jul;238:111280. doi: 10.1016/j.molbiopara.2020.111280. Epub 2020 May 12. Mol Biochem Parasitol. 2020. PMID: 32407750
-
A twinfilin-like protein coordinates karyokinesis by influencing mitotic spindle elongation and DNA replication in Leishmania.Mol Microbiol. 2016 Apr;100(1):173-87. doi: 10.1111/mmi.13310. Epub 2016 Feb 19. Mol Microbiol. 2016. PMID: 26713845
-
Emerging Functions of Actins and Actin Binding Proteins in Trypanosomatids.Front Cell Dev Biol. 2020 Oct 9;8:587685. doi: 10.3389/fcell.2020.587685. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33163497 Free PMC article. Review.
-
Profilin 1 plays feedback role in actin-mediated polar body extrusion in mouse oocytes.Reprod Fertil Dev. 2018 May;30(5):752-758. doi: 10.1071/RD17354. Reprod Fertil Dev. 2018. PMID: 29096761
-
Profilin, a multi-modal regulator of neuronal plasticity.Bioessays. 2008 Oct;30(10):994-1002. doi: 10.1002/bies.20822. Bioessays. 2008. PMID: 18798527 Review.
Cited by
-
Eukaryotic translation initiation factor 4A1 in the pathogenesis and treatment of cancers.Front Mol Biosci. 2023 Nov 9;10:1289650. doi: 10.3389/fmolb.2023.1289650. eCollection 2023. Front Mol Biosci. 2023. PMID: 38028556 Free PMC article. Review.
-
Analysis of the Leishmania mexicana promastigote cell cycle using imaging flow cytometry provides new insights into cell cycle flexibility and events of short duration.PLoS One. 2024 Oct 3;19(10):e0311367. doi: 10.1371/journal.pone.0311367. eCollection 2024. PLoS One. 2024. PMID: 39361666 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

