Concurrent CDX2 cis-deregulation and UBTF::ATXN7L3 fusion define a novel high-risk subtype of B-cell ALL
- PMID: 35316324
- PMCID: PMC9203705
- DOI: 10.1182/blood.2021014723
Concurrent CDX2 cis-deregulation and UBTF::ATXN7L3 fusion define a novel high-risk subtype of B-cell ALL
Abstract
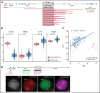
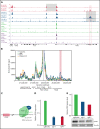
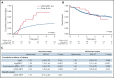
Oncogenic alterations underlying B-cell acute lymphoblastic leukemia (B-ALL) in adults remain incompletely elucidated. To uncover novel oncogenic drivers, we performed RNA sequencing and whole-genome analyses in a large cohort of unresolved B-ALL. We identified a novel subtype characterized by a distinct gene expression signature and the unique association of 2 genomic microdeletions. The 17q21.31 microdeletion resulted in a UBTF::ATXN7L3 fusion transcript encoding a chimeric protein. The 13q12.2 deletion resulted in monoallelic ectopic expression of the homeobox transcription factor CDX2, located 138 kb in cis from the deletion. Using 4C-sequencing and CRISPR interference experiments, we elucidated the mechanism of CDX2 cis-deregulation, involving PAN3 enhancer hijacking. CDX2/UBTF ALL (n = 26) harbored a distinct pattern of additional alterations including 1q gain and CXCR4 activating mutations. Within adult patients with Ph- B-ALL enrolled in GRAALL trials, patients with CDX2/UBTF ALL (n = 17/723, 2.4%) were young (median age, 31 years) and dramatically enriched in females (male/female ratio, 0.2, P = .002). They commonly presented with a pro-B phenotype ALL and moderate blast cell infiltration. They had poor response to treatment including a higher risk of failure to first induction course (19% vs 3%, P = .017) and higher post-induction minimal residual disease (MRD) levels (MRD ≥ 10-4, 93% vs 46%, P < .001). This early resistance to treatment translated into a significantly higher cumulative incidence of relapse (75.0% vs 32.4%, P = .004) in univariate and multivariate analyses. In conclusion, we discovered a novel B-ALL entity defined by the unique combination of CDX2 cis-deregulation and UBTF::ATXN7L3 fusion, representing a high-risk disease in young adults.
© 2022 by The American Society of Hematology.
Figures





Comment in
-
Why B(-)other? About the gap of unknowns in ALL.Blood. 2022 Jun 16;139(24):3455-3457. doi: 10.1182/blood.2022015993. Blood. 2022. PMID: 35708724 Free PMC article. No abstract available.
Similar articles
-
Enhancer retargeting of CDX2 and UBTF::ATXN7L3 define a subtype of high-risk B-progenitor acute lymphoblastic leukemia.Blood. 2022 Jun 16;139(24):3519-3531. doi: 10.1182/blood.2022015444. Blood. 2022. PMID: 35192684 Free PMC article.
-
UBTF::ATXN7L3 gene fusion defines novel B cell precursor ALL subtype with CDX2 expression and need for intensified treatment.Leukemia. 2022 Jun;36(6):1676-1680. doi: 10.1038/s41375-022-01557-6. Epub 2022 Apr 9. Leukemia. 2022. PMID: 35397658 Free PMC article. No abstract available.
-
Two novel high-risk adult B-cell acute lymphoblastic leukemia subtypes with high expression of CDX2 and IDH1/2 mutations.Blood. 2022 Mar 24;139(12):1850-1862. doi: 10.1182/blood.2021011921. Blood. 2022. PMID: 34695176
-
The clinical significance of CDX2 in leukemia: A new perspective for leukemia research.Leuk Res. 2018 Sep;72:45-51. doi: 10.1016/j.leukres.2018.07.021. Epub 2018 Jul 29. Leuk Res. 2018. PMID: 30096576 Review.
-
An update on PCR use for minimal residual disease monitoring in acute lymphoblastic leukemia.Expert Rev Mol Diagn. 2017 Nov;17(11):953-963. doi: 10.1080/14737159.2017.1377073. Epub 2017 Sep 21. Expert Rev Mol Diagn. 2017. PMID: 28891364 Review.
Cited by
-
Association of leukemic molecular profile with efficacy of inotuzumab ozogamicin in adults with relapsed/refractory ALL.Blood Adv. 2024 Jun 25;8(12):3226-3236. doi: 10.1182/bloodadvances.2023012430. Blood Adv. 2024. PMID: 38607410 Free PMC article. Clinical Trial.
-
Oncogenic lesions and molecular subtypes in adults with B-cell acute lymphoblastic leukemia.Cancer Sci. 2023 Jan;114(1):8-15. doi: 10.1111/cas.15583. Epub 2022 Sep 27. Cancer Sci. 2023. PMID: 36106363 Free PMC article. Review.
-
Why B(-)other? About the gap of unknowns in ALL.Blood. 2022 Jun 16;139(24):3455-3457. doi: 10.1182/blood.2022015993. Blood. 2022. PMID: 35708724 Free PMC article. No abstract available.
-
Acute myeloid leukemias with UBTF tandem duplications are sensitive to menin inhibitors.Blood. 2024 Feb 15;143(7):619-630. doi: 10.1182/blood.2023021359. Blood. 2024. PMID: 37890156 Free PMC article.
-
Acute lymphoblastic leukaemia.Nat Rev Dis Primers. 2024 Jun 13;10(1):41. doi: 10.1038/s41572-024-00525-x. Nat Rev Dis Primers. 2024. PMID: 38871740 Review.
References
-
- Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278(5340):1059-1064. - PubMed
-
- Yeoh E-J, Ross ME, Shurtleff SA, et al. . Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1(2):133-143. - PubMed
-
- Huguet F, Chevret S, Leguay T, et al. ; Group of Research on Adult ALL (GRAALL) . Intensified therapy of acute lymphoblastic leukemia in adults: report of the randomized GRAALL-2005 clinical trial. J Clin Oncol. 2018;36(24):2514-2523. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

