Strategies of preserving genetic diversity while maximizing genetic response from implementing genomic selection in pulse breeding programs
- PMID: 35316351
- PMCID: PMC9205836
- DOI: 10.1007/s00122-022-04071-6
Strategies of preserving genetic diversity while maximizing genetic response from implementing genomic selection in pulse breeding programs
Abstract
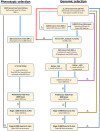
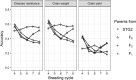
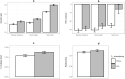
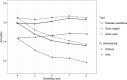
Genomic selection maximizes genetic gain by recycling parents to germplasm pool earlier and preserves genetic diversity by restricting the number of fixed alleles and the relationship in pulse breeding programs. Using a stochastic computer simulation, we investigated the benefit of optimization strategies in the context of genomic selection (GS) for pulse breeding programs. We simulated GS for moderately complex to highly complex traits such as disease resistance, grain weight and grain yield in multiple environments with a high level of genotype-by-environment interaction for grain yield. GS led to higher genetic gain per unit of time and higher genetic diversity loss than phenotypic selection by shortening the breeding cycle time. The genetic gain obtained from selecting the segregating parents early in the breeding cycle (at F1 or F2 stages) was substantially higher than selecting at later stages even though prediction accuracy was moderate. Increasing the number of F1 intercross (F1i) families and keeping the total number of progeny of F1i families constant, we observed a decrease in genetic gain and increase in genetic diversity, whereas increasing the number of progeny per F1i family while keeping a constant number of F1i families increased the rate of genetic gain and had higher genetic diversity loss per unit of time. Adding 50 F2 family phenotypes to the training population increased the accuracy of genomic breeding values (GEBVs) and genetic gain per year and decreased the rate of genetic diversity loss. Genetic diversity could be preserved by applying a strategy that restricted both the percentage of alleles fixed and the average relationship of the group of selected parents to preserve long-term genetic improvement in the pulse breeding program.
© 2022. The Author(s).
Conflict of interest statement
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Figures



Similar articles
-
Genetic Gain and Inbreeding from Genomic Selection in a Simulated Commercial Breeding Program for Perennial Ryegrass.Plant Genome. 2016 Mar;9(1). doi: 10.3835/plantgenome2015.06.0046. Plant Genome. 2016. PMID: 27898764
-
Gains through selection for grain yield in a winter wheat breeding program.PLoS One. 2020 Apr 28;15(4):e0221603. doi: 10.1371/journal.pone.0221603. eCollection 2020. PLoS One. 2020. PMID: 32343696 Free PMC article.
-
Optimal cross selection for long-term genetic gain in two-part programs with rapid recurrent genomic selection.Theor Appl Genet. 2018 Sep;131(9):1953-1966. doi: 10.1007/s00122-018-3125-3. Epub 2018 Jun 6. Theor Appl Genet. 2018. PMID: 29876589 Free PMC article.
-
Advances in integrated genomic selection for rapid genetic gain in crop improvement: a review.Planta. 2022 Sep 23;256(5):87. doi: 10.1007/s00425-022-03996-y. Planta. 2022. PMID: 36149531 Review.
-
Understanding the genomic selection for crop improvement: current progress and future prospects.Mol Genet Genomics. 2023 Jul;298(4):813-821. doi: 10.1007/s00438-023-02026-0. Epub 2023 May 10. Mol Genet Genomics. 2023. PMID: 37162565 Review.
Cited by
-
Genetic Gain and Inbreeding in Different Simulated Genomic Selection Schemes for Grain Yield and Oil Content in Safflower.Plants (Basel). 2024 Jun 6;13(11):1577. doi: 10.3390/plants13111577. Plants (Basel). 2024. PMID: 38891385 Free PMC article.
-
Harnessing Multi-Omics Strategies and Bioinformatics Innovations for Advancing Soybean Improvement: A Comprehensive Review.Plants (Basel). 2024 Sep 28;13(19):2714. doi: 10.3390/plants13192714. Plants (Basel). 2024. PMID: 39409584 Free PMC article. Review.
-
110 years of rice breeding at LSU: realized genetic gains and future optimization.Theor Appl Genet. 2025 Jun 9;138(7):142. doi: 10.1007/s00122-025-04913-z. Theor Appl Genet. 2025. PMID: 40488752 Free PMC article.
-
Genomic-inferred cross-selection methods for multi-trait improvement in a recurrent selection breeding program.Plant Methods. 2024 Sep 2;20(1):133. doi: 10.1186/s13007-024-01258-4. Plant Methods. 2024. PMID: 39218896 Free PMC article.
-
Optimizing the selection of quantitative traits in plant breeding using simulation.Front Plant Sci. 2025 Feb 10;16:1495662. doi: 10.3389/fpls.2025.1495662. eCollection 2025. Front Plant Sci. 2025. PMID: 39996117 Free PMC article. Review.
References
-
- Bernardo R, Yu J. Prospects for genomewide selection for quantitative traits in maize. Crop Sci. 2007;47:1082–1090. doi: 10.2135/cropsci2006.11.0690. - DOI
-
- Bhandari H, Bhanu AN, Srivastava K, Singh M, Shreya HA. Assessment of genetic diversity in crop plants—an overview. Adv Plants Agric Res. 2017;7:279–286.
-
- Brisbane JR, Gibson JP. Balancing selection response and inbreeding by including predicted stabilised genetic contributions in selection decisions. GSE. 1995;27:541. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

