Determining the Effects of Neddylation on Cullin-RING Ligase-Dependent Protein Ubiquitination
- PMID: 35316580
- PMCID: PMC8969890
- DOI: 10.1002/cpz1.401
Determining the Effects of Neddylation on Cullin-RING Ligase-Dependent Protein Ubiquitination
Abstract
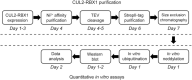
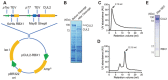
As the largest family of ubiquitin (Ub) E3 ligases, cullin-RING ligases (CRLs) play crucial roles in various cellular processes, and their activities are tightly regulated by orchestrated mechanisms. Neddylation, the conjugation of a Ub-like protein NEDD8 to a target protein such as the cullin, represents a key regulatory mechanism for CRLs. Biochemical and structural studies of a few CRLs have revealed that cullin neddylation alters the CRL conformation and activates CRL-dependent protein ubiquitination. Here, using CUL2-RING ligase (CRL2) as an example, we describe our protocols for the preparation of recombinant CUL2 with or without NEDD8 conjugation, which is further used to quantitatively determine the effect of neddylation on CRL2-dependent protein ubiquitination in vitro. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC. Basic Protocol 1: Expression and purification of CUL2•RBX1 from Escherichia coli Support Protocol: Further purification of CUL2•RBX1 with additional chromatography on an FPLC system Basic Protocol 2: Reconstitution of cullin neddylation for quantitative ubiquitination assay in vitro.
Keywords: cullin-RING ligases; in vitro assay; neddylation; protein purification; ubiquitination.
© 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Modulation of Cullin-RING E3 ubiquitin ligase-dependent ubiquitination by small molecule compounds.J Biol Chem. 2024 Mar;300(3):105752. doi: 10.1016/j.jbc.2024.105752. Epub 2024 Feb 13. J Biol Chem. 2024. PMID: 38354780 Free PMC article.
-
Neddylation-induced conformational control regulates cullin RING ligase activity in vivo.J Mol Biol. 2011 Jun 3;409(2):136-45. doi: 10.1016/j.jmb.2011.03.023. Epub 2011 Apr 2. J Mol Biol. 2011. PMID: 21463634
-
Introduction.Adv Exp Med Biol. 2020;1217:1-8. doi: 10.1007/978-981-15-1025-0_1. Adv Exp Med Biol. 2020. PMID: 31898218
-
Cullin-RING E3 Ubiquitin Ligases: Bridges to Destruction.Subcell Biochem. 2017;83:323-347. doi: 10.1007/978-3-319-46503-6_12. Subcell Biochem. 2017. PMID: 28271482 Free PMC article. Review.
-
Assembly and Regulation of CRL Ubiquitin Ligases.Adv Exp Med Biol. 2020;1217:33-46. doi: 10.1007/978-981-15-1025-0_3. Adv Exp Med Biol. 2020. PMID: 31898220 Review.
Cited by
-
Identification of Potential Neddylation-related Key Genes in Ischemic Stroke based on Machine Learning Methods.Mol Neurobiol. 2024 May;61(5):2530-2541. doi: 10.1007/s12035-023-03738-5. Epub 2023 Nov 1. Mol Neurobiol. 2024. PMID: 37910287
-
Molecular mechanisms of CAND2 in regulating SCF ubiquitin ligases.Nat Commun. 2025 Feb 26;16(1):1998. doi: 10.1038/s41467-025-57065-5. Nat Commun. 2025. PMID: 40011427 Free PMC article.
-
Review: A silent concert in developing plants: Dynamic assembly of cullin-RING ubiquitin ligases.Plant Sci. 2023 May;330:111662. doi: 10.1016/j.plantsci.2023.111662. Epub 2023 Feb 22. Plant Sci. 2023. PMID: 36822503 Free PMC article. Review.
-
CAND1 inhibits Cullin-2-RING ubiquitin ligases for enhanced substrate specificity.Nat Struct Mol Biol. 2024 Feb;31(2):323-335. doi: 10.1038/s41594-023-01167-5. Epub 2024 Jan 4. Nat Struct Mol Biol. 2024. PMID: 38177676 Free PMC article.
-
Ubiquitination in lipid metabolism reprogramming: implications for pediatric solid tumors.Front Immunol. 2025 Apr 30;16:1554311. doi: 10.3389/fimmu.2025.1554311. eCollection 2025. Front Immunol. 2025. PMID: 40370434 Free PMC article. Review.
References
-
- Davarinejad, H. (2015). Quantifications of western blots with ImageJ. University of York. Retrieved from http://www.yorku.ca/yisheng/Internal/Protocols/ImageJ.pdf.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

