Phages and their satellites encode hotspots of antiviral systems
- PMID: 35316646
- PMCID: PMC9122126
- DOI: 10.1016/j.chom.2022.02.018
Phages and their satellites encode hotspots of antiviral systems
Abstract
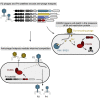
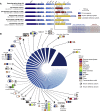
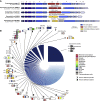
Bacteria carry diverse genetic systems to defend against viral infection, some of which are found within prophages where they inhibit competing viruses. Phage satellites pose additional pressures on phages by hijacking key viral elements to their own benefit. Here, we show that E. coli P2-like phages and their parasitic P4-like satellites carry hotspots of genetic variation containing reservoirs of anti-phage systems. We validate the activity of diverse systems and describe PARIS, an abortive infection system triggered by a phage-encoded anti-restriction protein. Antiviral hotspots participate in inter-viral competition and shape dynamics between the bacterial host, P2-like phages, and P4-like satellites. Notably, the anti-phage activity of satellites can benefit the helper phage during competition with virulent phages, turning a parasitic relationship into a mutualistic one. Anti-phage hotspots are present across distant species and constitute a substantial source of systems that participate in the competition between mobile genetic elements.
Keywords: abortive infection; bacterial immunity; genetic diversity; inter-viral competition; mobile genetic elements; phage defense; phage satellite.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.B. is a founder of Eligo Bioscience and a member of its scientific advisory board.
Figures





References
-
- Beilstein F., Dreiseikelmann B. Temperate bacteriophage ΦO18P from an Aeromonas media isolate: characterization and complete genome sequence. Virology. 2008;373:25–29. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

