N-Acetyldopamine dimers from Oxya chinensis sinuosa attenuates lipopolysaccharides induced inflammation and inhibits cathepsin C activity
- PMID: 35317232
- PMCID: PMC8908036
- DOI: 10.1016/j.csbj.2022.02.011
N-Acetyldopamine dimers from Oxya chinensis sinuosa attenuates lipopolysaccharides induced inflammation and inhibits cathepsin C activity
Abstract
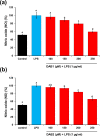
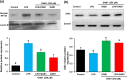
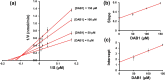
Oxya chinensis sinuosa (rice field grasshopper) is an edible insect with numerous health beneficial properties, traditionally being used to treat many ailments in Korea and other countries. O. chinensis sinuosa has been used from centuries, however, a little is known about the chemical functionality of its bioactive compounds. Therefore, this study examined the anti-inflammatory and cathepsin C inhibitory activities of N-acetyldopamine dimer (2R, 3S)-2-(3',4'-dihydroxyphenyl)-3-acetylamino-7-(N-acetyl-2″-aminoethyl)-1,4-benzodioxane (DAB1) isolated from O. chinensis sinuosa. Results showed that DAB1 reduced the expression of pro-inflammatory mediator (iNOS, COX-2) and cytokines (TNF-α, IL-1β, and IL-6), and curtailed the nuclear translocation of NF-κB by inhibiting the phosphorylation of IκBα in lipopolysaccharide stimulated macrophages. Additionally, DAB1 inhibited cathepsin C activity at the cellular level, supported by in vitro assay (Ki, 71.56 ± 10.21 µM and Kis, 133.55 ± 18.2 µM). Moreover, combinatorial molecular simulation and binding free energy analysis suggested a significant stability and binding affinity of cathepsin C-DAB1 complex via formation of hydrogen bond and hydrophobic interactions with the catalytic residues (Gln228, Thr379, Asn380, and Hie381). Also, essential dynamics analysis showed DAB1 induced non-functional motions in cathepsin C structure. Collectively, DAB1 was concluded as anti-inflammatory and cathepsin C inhibiting agent and could be used in the drug development against respective diseases.
Keywords: Cathepsin C; Essential dynamics; In silico; Inflammation; NF-κB; Oxya chinensis sinuosa.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
-
- Nathan C. Points of control in inflammation. Nature. 2002;420(6917):846–852. - PubMed
-
- Wan P., Xie M., Chen G., Dai Z., Hu B., Zeng X., et al. Anti-inflammatory effects of dicaffeoylquinic acids from Ilex kudingcha on lipopolysaccharide-treated RAW264.7 macrophages and potential mechanisms. Food Chem Toxicol. 2019;126:332–342. - PubMed
-
- Tasneem S., Liu B., Li B., Choudhary M.I., Wang W. Molecular pharmacology of inflammation: Medicinal plants as anti-inflammatory agents. Pharmacol Res. 2019;139:126–140. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
