Identification of piRNA disease associations using deep learning
- PMID: 35317234
- PMCID: PMC8908038
- DOI: 10.1016/j.csbj.2022.02.026
Identification of piRNA disease associations using deep learning
Abstract
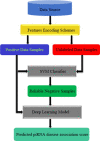
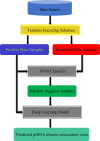
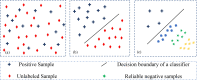
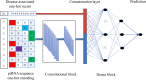
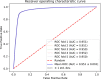
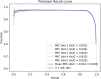
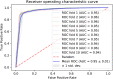
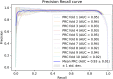
Piwi-interacting RNAs (piRNAs) play a pivotal role in maintaining genome integrity by repression of transposable elements, gene stability, and association with various disease progressions. Cost-efficient computational methods for the identification of piRNA disease associations promote the efficacy of disease-specific drug development. In this regard, we developed a simple, robust, and efficient deep learning method for identifying the piRNA disease associations known as piRDA. The proposed architecture extracts the most significant and abstract information from raw sequences represented in a simplicated piRNA disease pair without any involvement of features engineering. Two-step positive unlabeled learning and bootstrapping technique are utilized to abstain from the false-negative and biased predictions dealing with positive unlabeled data. The performance of proposed method piRDA is evaluated using k-fold cross-validation. The piRDA is significantly improved in all the performance evaluation measures for the identification of piRNA disease associations in comparison to state-of-the-art method. Moreover, it is thus projected conclusively that the proposed computational method could play a significant role as a supportive and practical tool for primitive disease mechanisms and pharmaceutical research such as in academia and drug design. Eventually, the proposed model can be accessed using publicly available and user-friendly web tool athttp://nsclbio.jbnu.ac.kr/tools/piRDA/.
Keywords: Convolutional Neural Network; Deep learning; Positive unlabeled learning; Reliable negative sample; Sequence analysis; Web-server; piRNA disease associations.
© 2022 The Authors. Published by Elsevier B.V. on behalf of Research Network of Computational and Structural Biotechnology.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
PUTransGCN: identification of piRNA-disease associations based on attention encoding graph convolutional network and positive unlabelled learning.Brief Bioinform. 2024 Mar 27;25(3):bbae144. doi: 10.1093/bib/bbae144. Brief Bioinform. 2024. PMID: 38581419 Free PMC article.
-
Identification of Functional piRNAs Using a Convolutional Neural Network.IEEE/ACM Trans Comput Biol Bioinform. 2022 May-Jun;19(3):1661-1669. doi: 10.1109/TCBB.2020.3034313. Epub 2022 Jun 3. IEEE/ACM Trans Comput Biol Bioinform. 2022. PMID: 33119510
-
iPiDA-sHN: Identification of Piwi-interacting RNA-disease associations by selecting high quality negative samples.Comput Biol Chem. 2020 Oct;88:107361. doi: 10.1016/j.compbiolchem.2020.107361. Epub 2020 Aug 29. Comput Biol Chem. 2020. PMID: 32916452
-
Databases and computational methods for the identification of piRNA-related molecules: A survey.Comput Struct Biotechnol J. 2024 Jan 22;23:813-833. doi: 10.1016/j.csbj.2024.01.011. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38328006 Free PMC article. Review.
-
PiWi RNA in Neurodevelopment and Neurodegenerative Disorders.Curr Mol Pharmacol. 2022;15(3):517-531. doi: 10.2174/1874467214666210629164535. Curr Mol Pharmacol. 2022. PMID: 34212832 Review.
Cited by
-
Revealing differential expression patterns of piRNA in FACS blood cells of SARS-CoV-2 infected patients.BMC Med Genomics. 2024 Aug 14;17(1):212. doi: 10.1186/s12920-024-01982-9. BMC Med Genomics. 2024. PMID: 39143590 Free PMC article.
-
iPiDA-GCN: Identification of piRNA-disease associations based on Graph Convolutional Network.PLoS Comput Biol. 2022 Oct 27;18(10):e1010671. doi: 10.1371/journal.pcbi.1010671. eCollection 2022 Oct. PLoS Comput Biol. 2022. PMID: 36301998 Free PMC article.
-
PUTransGCN: identification of piRNA-disease associations based on attention encoding graph convolutional network and positive unlabelled learning.Brief Bioinform. 2024 Mar 27;25(3):bbae144. doi: 10.1093/bib/bbae144. Brief Bioinform. 2024. PMID: 38581419 Free PMC article.
-
iPiDA-SWGCN: Identification of piRNA-disease associations based on Supplementarily Weighted Graph Convolutional Network.PLoS Comput Biol. 2023 Jun 20;19(6):e1011242. doi: 10.1371/journal.pcbi.1011242. eCollection 2023 Jun. PLoS Comput Biol. 2023. PMID: 37339125 Free PMC article.
-
piOxi database: a web resource of germline and somatic tissue piRNAs identified by chemical oxidation.Database (Oxford). 2024 Jan 10;2024:baad096. doi: 10.1093/database/baad096. Database (Oxford). 2024. PMID: 38204359 Free PMC article.
References
-
- D. Meseure, K.D. Alsibai, Part 1: The piwi-pirna pathway is an immune-like surveillance process that controls genome integrity by silencing transposable elements, in: Chromatin and Epigenetics, IntechOpen, 2018.
-
- Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small rnas binds mammalian piwi proteins. Nature. 2006;442(7099):199–202. - PubMed
-
- Sasaki T., Shiohama A., Minoshima S., Shimizu N. Identification of eight members of the argonaute family in the human genome. Genomics. 2003;82(3):323–330. - PubMed
LinkOut - more resources
Full Text Sources
