Exploring a prolonged enterovirus C104 infection in a severely ill patient using nanopore sequencing
- PMID: 35317350
- PMCID: PMC8932292
- DOI: 10.1093/ve/veab109
Exploring a prolonged enterovirus C104 infection in a severely ill patient using nanopore sequencing
Abstract
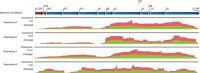
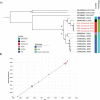
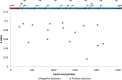
Chronic enterovirus infections can cause significant morbidity, particularly in immunocompromised patients. This study describes a fatal case associated with a chronic untypeable enterovirus infection in an immunocompromised patient admitted to a Dutch university hospital over nine months. We aimed to identify the enterovirus genotype responsible for the infection and to determine potential evolutionary changes. Long-read sequencing was performed using viral targeted sequence capture on four respiratory and one faecal sample. Phylogenetic analysis was performed using a maximum likelihood method, along with a root-to-tip regression and time-scaled phylogenetic analysis to estimate evolutionary changes between sample dates. Intra-host variant detection, using a Fixed Ploidy algorithm, and selection pressure, using a Fixed Effect Likelihood and a Mixed Effects Model of Evolution, were also used to explore the patient samples. Near-complete genomes of enterovirus C104 (EV-C104) were recovered in all respiratory samples but not in the faecal sample. The recovered genomes clustered with a recently reported EV-C104 from Belgium in August 2018. Phylodynamic analysis including ten available EV-C104 genomes, along with the patient sequences, estimated the most recent common ancestor to occur in the middle of 2005 with an overall estimated evolution rate of 2.97 × 10-3 substitutions per year. Although positive selection pressure was identified in the EV-C104 reference sequences, the genomes recovered from the patient samples alone showed an overall negative selection pressure in multiple codon sites along the genome. A chronic infection resulting in respiratory failure from a relatively rare enterovirus was observed in a transplant recipient. We observed an increase in single-nucleotide variations between sample dates from a rapidly declining patient, suggesting mutations are weakly deleterious and have not been purged during selection. This is further supported by the persistence of EV-C104 in the patient, despite the clearance of other viral infections. Next-generation sequencing with viral enrichment could be used to detect and characterise challenging samples when conventional workflows are insufficient.
Keywords: chronic infection; enterovirus; immunocompromised; intra-host evolution; nanopore sequencing; virus evolution.
© The Author(s) 2022. Published by Oxford University Press.
Figures

References
LinkOut - more resources
Full Text Sources

