Atrophic gastritis and gastric cancer tissue miRNome analysis reveals hsa-miR-129-1 and hsa-miR-196a as potential early diagnostic biomarkers
- PMID: 35317427
- PMCID: PMC8900545
- DOI: 10.3748/wjg.v28.i6.653
Atrophic gastritis and gastric cancer tissue miRNome analysis reveals hsa-miR-129-1 and hsa-miR-196a as potential early diagnostic biomarkers
Abstract
Background: Gastric cancer (GC) is one of the most frequently diagnosed tumor globally. In most cases, GC develops in a stepwise manner from chronic gastritis or atrophic gastritis (AG) to cancer. One of the major issues in clinical settings of GC is diagnosis at advanced disease stages resulting in poor prognosis. MicroRNAs (miRNAs) are small noncoding molecules that play an essential role in a variety of fundamental biological processes. However, clinical potential of miRNA profiling in the gastric cancerogenesis, especially in premalignant GC cases, remains unclear.
Aim: To evaluate the AG and GC tissue miRNomes and identify specific miRNAs' potential for clinical applications (e.g., non-invasive diagnostics).
Methods: Study included a total of 125 subjects: Controls (CON), AG, and GC patients. All study subjects were recruited at the Departments of Surgery or Gastroenterology, Hospital of Lithuanian University of Health Sciences and divided into the profiling (n = 60) and validation (n = 65) cohorts. Total RNA isolated from tissue samples was used for preparation of small RNA sequencing libraries and profiled using next-generation sequencing (NGS). Based on NGS data, deregulated miRNAs hsa-miR-129-1-3p and hsa-miR-196a-5p were analyzed in plasma samples of independent cohort consisting of CON, AG, and GC patients. Expression level of hsa-miR-129-1-3p and hsa-miR-196a-5p was determined using the quantitative real-time polymerase chain reaction and 2-ΔΔCt method.
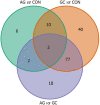
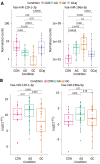
Results: Results of tissue analysis revealed 20 differentially expressed miRNAs in AG group compared to CON group, 129 deregulated miRNAs in GC compared to CON, and 99 altered miRNAs comparing GC and AG groups. Only 2 miRNAs (hsa-miR-129-1-3p and hsa-miR-196a-5p) were identified to be step-wise deregulated in healthy-premalignant-malignant sequence. Area under the curve (AUC)-receiver operating characteristic analysis revealed that expression level of hsa-miR-196a-5p is significant for discrimination of CON vs AG, CON vs GC and AG vs GC and resulted in AUCs: 88.0%, 93.1% and 66.3%, respectively. Compar-ing results in tissue and plasma samples, hsa-miR-129-1-3p was significantly down-regulated in GC compared to AG (P = 0.0021 and P = 0.024, tissue and plasma, respectively). Moreover, analysis revealed that hsa-miR-215-3p/5p and hsa-miR-934 were significantly deregulated in GC based on Helicobacter pylori (H. pylori) infection status [log2 fold change (FC) = -4.52, P-adjusted = 0.02; log2FC = -4.00, P-adjusted = 0.02; log2FC = 6.09, P-adjusted = 0.02, respectively].
Conclusion: Comprehensive miRNome study provides evidence for gradual deregulation of hsa-miR-196a-5p and hsa-miR-129-1-3p in gastric carcinogenesis and found hsa-miR-215-3p/5p and hsa-miR-934 to be significantly deregulated in H. pylori carrying GC patients.
Keywords: Atrophic gastritis; Biomarkers; Gastric cancer; Helicobacter pylori; MicroRNAs; Tumorigenesis.
©The Author(s) 2022. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: The authors have declared no conflicts of interest.
Figures


References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol. 2014;11:664–674. - PubMed
-
- Tan P, Yeoh KG. Genetics and Molecular Pathogenesis of Gastric Adenocarcinoma. Gastroenterology. 2015;149:1153–1162.e3. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

