CIP4 targeted to recruit GTP-Cdc42 involving in invadopodia formation via NF-κB signaling pathway promotes invasion and metastasis of CRC
- PMID: 35317515
- PMCID: PMC8924540
- DOI: 10.1016/j.omto.2022.02.023
CIP4 targeted to recruit GTP-Cdc42 involving in invadopodia formation via NF-κB signaling pathway promotes invasion and metastasis of CRC
Abstract
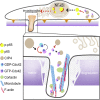
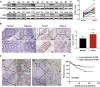
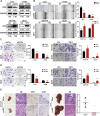
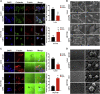
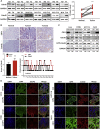
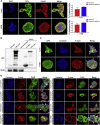
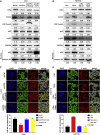
Cdc42-interacting protein 4 (CIP4), a member of the F-BAR family, which plays an important role in regulating cell membrane and actin, has been reported to interact with Cdc42 and be closely associated with tumor invadopodia formation. In this study, we found that CIP4 expression was significantly higher in human CRC tissues and correlated with the CRC infiltrating depth and metastasis, as well as the lower survival rate in patients. In cultured CRC cells, knockdown of CIP4 inhibited cell migration and invasion ability in vitro and tumor metastasis in vivo, while the overexpression of CIP4 promoted invadopodia formation and matrix degradation ability. We then identified GTP-Cdc42 as a directly interactive protein of CIP4, which was upregulated and recruited by CIP4. Furthermore, activated NF-κB signaling pathway was found in CIP4 overexpression of CRC cells contributing to invadopodia formation, while the inhibition of either CIP4 or Cdc42 led to the suppression of the NF-κB pathway and resulted in a decreased quantity of invadopodia. Our findings suggested that CIP4 targets to recruit GTP-Cdc42 and directly combines with it to accelerate invadopodia formation and function by activating NF-κB signaling pathway, thus promoting CRC infiltration and metastasis.
Keywords: CIP4; Cdc42; NF-κB; colorectal cancer; invadopodia; invasion; metastasis.
© 2022 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Cdc42-interacting protein 4 is a Src substrate that regulates invadopodia and invasiveness of breast tumors by promoting MT1-MMP endocytosis.J Cell Sci. 2011 May 15;124(Pt 10):1739-51. doi: 10.1242/jcs.078014. Epub 2011 Apr 26. J Cell Sci. 2011. PMID: 21525036
-
CDC42-interacting protein 4 promotes metastasis of nasopharyngeal carcinoma by mediating invadopodia formation and activating EGFR signaling.J Exp Clin Cancer Res. 2017 Jan 28;36(1):21. doi: 10.1186/s13046-016-0483-z. J Exp Clin Cancer Res. 2017. PMID: 28129778 Free PMC article.
-
AKAP-9 promotes colorectal cancer development by regulating Cdc42 interacting protein 4.Biochim Biophys Acta. 2016 Jun;1862(6):1172-81. doi: 10.1016/j.bbadis.2016.03.012. Epub 2016 Apr 12. Biochim Biophys Acta. 2016. PMID: 27039663 Free PMC article.
-
Identification of a CIP4 PKA phosphorylation site involved in the regulation of cancer cell invasiveness and metastasis.Cancer Lett. 2019 Oct 1;461:65-77. doi: 10.1016/j.canlet.2019.07.006. Epub 2019 Jul 15. Cancer Lett. 2019. PMID: 31319138
-
CIP4 coordinates with phospholipids and actin-associated proteins to localize to the protruding edge and produce actin ribs and veils.J Cell Sci. 2013 Jun 1;126(Pt 11):2411-23. doi: 10.1242/jcs.117473. Epub 2013 Apr 9. J Cell Sci. 2013. PMID: 23572514 Free PMC article.
Cited by
-
RPL21 interacts with LAMP3 to promote colorectal cancer invasion and metastasis by regulating focal adhesion formation.Cell Mol Biol Lett. 2023 Apr 16;28(1):31. doi: 10.1186/s11658-023-00443-y. Cell Mol Biol Lett. 2023. PMID: 37062845 Free PMC article.
-
miR-424-5p Promotes Proliferation, Migration and Invasion of Colorectal Cancer Cells via the Targeting TXNIP/Hippo Axi.Int J Gen Med. 2025 Jan 16;18:261-271. doi: 10.2147/IJGM.S497401. eCollection 2025. Int J Gen Med. 2025. PMID: 39834913 Free PMC article.
-
SLC38A3 Promotes the Proliferation and Migration of Tumor Cells and Predicts Poor Prognosis in Colorectal Cancer.ACS Omega. 2024 May 2;9(19):21116-21126. doi: 10.1021/acsomega.4c00901. eCollection 2024 May 14. ACS Omega. 2024. PMID: 38764627 Free PMC article.
-
Lysosomes and LAMPs as Autophagy Drivers of Drug Resistance in Colorectal Cancer.Cells. 2025 Apr 11;14(8):574. doi: 10.3390/cells14080574. Cells. 2025. PMID: 40277899 Free PMC article. Review.
-
Tethered Exosomes Containing the Matrix Metalloproteinase MT1-MMP Contribute to Extracellular Matrix Degradation.J Extracell Vesicles. 2025 Jul;14(7):e70122. doi: 10.1002/jev2.70122. J Extracell Vesicles. 2025. PMID: 40704538 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

