UPF1 contributes to the maintenance of endometrial cancer stem cell phenotype by stabilizing LINC00963
- PMID: 35318304
- PMCID: PMC8940903
- DOI: 10.1038/s41419-022-04707-x
UPF1 contributes to the maintenance of endometrial cancer stem cell phenotype by stabilizing LINC00963
Abstract
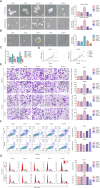
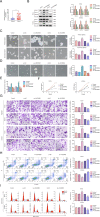
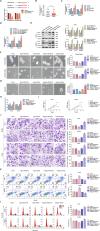
Endometrial cancer stem cells (ECSCs) play a vital role in endometrial cancer (EC) metastasis, relapse, and chemoresistance. However, the molecular mechanisms that sustain ECSCs remain elusive. Here, we showed that the expression of UPF1 was upregulated in EC tissues and ECSCs and correlated with poor clinicopathological characteristics. UPF1 silencing suppressed ECSC hallmarks, such as sphere formation ability, carboplatin resistance, migration and invasion, and cell cycle progression. UPF1 regulated the behavior and fate of ECSCs by stabilizing LINC00963. LINC00963 further shares the same miRNA response element with the core transcription factor SOX2 and relieved the suppression of SOX2 by miR-508-5p in self-renewing ECSCs. Notably, inhibition of UPF1 and LINC00963 in combination severely impaired the in vivo tumorigenic potential of ECSCs. We demonstrate that the UPF1/LINC00963/miR-508-5p/SOX2 axis has potential value in modulating ECSC maintenance, chemoresistance, and tumorigenesis in EC, which highlights a novel promising target for EC treatment.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
WTAP/IGF2BP3 mediated m6A modification of the EGR1/PTEN axis regulates the malignant phenotypes of endometrial cancer stem cells.J Exp Clin Cancer Res. 2024 Jul 23;43(1):204. doi: 10.1186/s13046-024-03120-w. J Exp Clin Cancer Res. 2024. PMID: 39044249 Free PMC article.
-
PVT1/miR-136/Sox2/UPF1 axis regulates the malignant phenotypes of endometrial cancer stem cells.Cell Death Dis. 2023 Mar 3;14(3):177. doi: 10.1038/s41419-023-05651-0. Cell Death Dis. 2023. PMID: 36869031 Free PMC article.
-
UPF1 impacts on mTOR signaling pathway and autophagy in endometrioid endometrial carcinoma.Aging (Albany NY). 2021 Sep 14;13(17):21202-21215. doi: 10.18632/aging.203421. Epub 2021 Sep 14. Aging (Albany NY). 2021. PMID: 34520393 Free PMC article.
-
Circ_0002577/miR-126-5p/MACC1 axis promotes endometrial carcinoma progression by regulation of proliferation, migration, invasion, and apoptosis of endometrial carcinoma cells.Arch Gynecol Obstet. 2022 Aug;306(2):481-491. doi: 10.1007/s00404-022-06412-8. Epub 2022 Feb 1. Arch Gynecol Obstet. 2022. PMID: 35103833
-
Bone Marrow-Derived Cells in Endometrial Cancer Pathogenesis: Insights from Breast Cancer.Cells. 2022 Feb 17;11(4):714. doi: 10.3390/cells11040714. Cells. 2022. PMID: 35203363 Free PMC article. Review.
Cited by
-
WTAP/IGF2BP3 mediated m6A modification of the EGR1/PTEN axis regulates the malignant phenotypes of endometrial cancer stem cells.J Exp Clin Cancer Res. 2024 Jul 23;43(1):204. doi: 10.1186/s13046-024-03120-w. J Exp Clin Cancer Res. 2024. PMID: 39044249 Free PMC article.
-
M2 macrophage-secreted KYNU promotes stemness remodeling and malignant behavior in endometrial cancer via the SOD2-mtROS-ERO1α-UPRER axis.J Exp Clin Cancer Res. 2025 Jul 4;44(1):193. doi: 10.1186/s13046-025-03285-y. J Exp Clin Cancer Res. 2025. PMID: 40616124 Free PMC article.
-
Engineered exosomes restore miR-508-5p expression in uterine corpus endometrial carcinoma and reduce tumor progression and metastasis by targeting DLL3.Front Oncol. 2025 Feb 24;15:1532564. doi: 10.3389/fonc.2025.1532564. eCollection 2025. Front Oncol. 2025. PMID: 40066092 Free PMC article.
-
LncRNA PRKCA-AS1 promotes LUAD progression and function as a ceRNA to regulate S100A16 by sponging miR-508-5p.J Cancer. 2024 Jan 27;15(6):1718-1730. doi: 10.7150/jca.91184. eCollection 2024. J Cancer. 2024. PMID: 38370382 Free PMC article.
-
ZNF432 suppresses endometrial cancer progression by promoting UPF1 ubiquitination and inducing apoptosis.Discov Oncol. 2025 Jun 14;16(1):1095. doi: 10.1007/s12672-025-02915-3. Discov Oncol. 2025. PMID: 40515959 Free PMC article.
References
-
- Wortman BG, Bosse T, Nout RA, Lutgens L, van der Steen-Banasik EM, Westerveld H, et al. Molecular-integrated risk profile to determine adjuvant radiotherapy in endometrial cancer: Evaluation of the pilot phase of the PORTEC-4a trial. Gynecol Oncol. 2018;151:69–75. - PubMed
-
- Emons G, Kim JW, Weide K, de Gregorio N, Wimberger P, Trillsch F, et al. Endometrial Cancer Lymphadenectomy Trial (ECLAT) (pelvic and para-aortic lymphadenectomy in patients with stage I or II endometrial cancer with high risk of recurrence; AGO-OP.6) Int J Gynecol Cancer. 2021;31:1075–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

