Structures of pseudorabies virus capsids
- PMID: 35318331
- PMCID: PMC8940892
- DOI: 10.1038/s41467-022-29250-3
Structures of pseudorabies virus capsids
Abstract
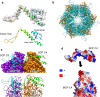
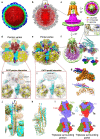
Pseudorabies virus (PRV) is a major etiological agent of swine infectious diseases and is responsible for significant economic losses in the swine industry. Recent data points to human viral encephalitis caused by PRV infection, suggesting that PRV may be able to overcome the species barrier to infect humans. To date, there is no available therapeutic for PRV infection. Here, we report the near-atomic structures of the PRV A-capsid and C-capsid, and illustrate the interaction that occurs between these subunits. We show that the C-capsid portal complex is decorated with capsid-associated tegument complexes. The PRV capsid structure is highly reminiscent of other α-herpesviruses, with some additional structural features of β- and γ-herpesviruses. These results illustrate the structure of the PRV capsid and elucidate the underlying assembly mechanism at the molecular level. This knowledge may be useful for the development of oncolytic agents or specific therapeutics against this arm of the herpesvirus family.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins.J Mol Biol. 2013 Sep 23;425(18):3415-28. doi: 10.1016/j.jmb.2013.06.034. Epub 2013 Jul 1. J Mol Biol. 2013. PMID: 23827137 Free PMC article.
-
Dissecting the Herpesvirus Architecture by Targeted Proteolysis.J Virol. 2018 Aug 16;92(17):e00738-18. doi: 10.1128/JVI.00738-18. Print 2018 Sep 1. J Virol. 2018. PMID: 29899099 Free PMC article.
-
CRISPR/Cas9-Constructed Pseudorabies Virus Mutants Reveal the Importance of UL13 in Alphaherpesvirus Escape from Genome Silencing.J Virol. 2021 Feb 24;95(6):e02286-20. doi: 10.1128/JVI.02286-20. Print 2021 Feb 24. J Virol. 2021. PMID: 33361431 Free PMC article.
-
Membrane fusion, potential threats, and natural antiviral drugs of pseudorabies virus.Vet Res. 2023 May 2;54(1):39. doi: 10.1186/s13567-023-01171-z. Vet Res. 2023. PMID: 37131259 Free PMC article. Review.
-
Current Status and Challenge of Pseudorabies Virus Infection in China.Virol Sin. 2021 Aug;36(4):588-607. doi: 10.1007/s12250-020-00340-0. Epub 2021 Feb 22. Virol Sin. 2021. PMID: 33616892 Free PMC article. Review.
Cited by
-
Identification, Pathogenicity, and Reverse Genetics System Construction of a Pseudorabies Virus Isolate from Pigs in China.Vet Sci. 2025 May 26;12(6):519. doi: 10.3390/vetsci12060519. Vet Sci. 2025. PMID: 40559756 Free PMC article.
-
The 25-kDa linear polyethylenimine exerts specific antiviral activity against pseudorabies virus through interferencing its adsorption via electrostatic interaction.J Virol. 2024 Mar 19;98(3):e0000724. doi: 10.1128/jvi.00007-24. Epub 2024 Feb 2. J Virol. 2024. PMID: 38305153 Free PMC article.
-
Nanomaterial Adjuvants for Veterinary Vaccines: Mechanisms and Applications.Research (Wash D C). 2025 Jul 8;8:0761. doi: 10.34133/research.0761. eCollection 2025. Research (Wash D C). 2025. PMID: 40636132 Free PMC article. Review.
-
Pseudorabies virus VHS protein abrogates interferon responses by blocking NF-κB and IRF3 nuclear translocation.Virol Sin. 2024 Aug;39(4):587-599. doi: 10.1016/j.virs.2024.05.009. Epub 2024 May 30. Virol Sin. 2024. PMID: 38823782 Free PMC article.
-
Enhanced Anti-Tumor Response Elicited by a Novel Oncolytic Pseudorabies Virus Engineered with a PD-L1 Inhibitor.Viruses. 2024 Jul 31;16(8):1228. doi: 10.3390/v16081228. Viruses. 2024. PMID: 39205202 Free PMC article.
References
-
- Mettenleiter TC. Molecular biology of pseudorabies (Aujeszky’s disease) virus. Comp. Immunol., Microbiol. Infect. Dis. 1991;14:151–163. - PubMed
-
- Skinner G, Ahmad A, Davies JA. The infrequency of transmission of herpesviruses between humans and animals; postulation of an unrecognised protective host mechanism. Comp. Immunol. Microbiol. Infect. Dis. 2001;24:255–269. - PubMed
-
- Freuling CM, Muller TF, Mettenleiter TC. Vaccines against pseudorabies virus (PrV) Vet. Microbiol. 2017;206:3–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

