A deep-learning approach for online cell identification and trace extraction in functional two-photon calcium imaging
- PMID: 35318335
- PMCID: PMC8940911
- DOI: 10.1038/s41467-022-29180-0
A deep-learning approach for online cell identification and trace extraction in functional two-photon calcium imaging
Abstract
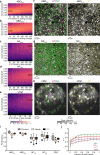
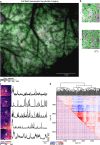
In vivo two-photon calcium imaging is a powerful approach in neuroscience. However, processing two-photon calcium imaging data is computationally intensive and time-consuming, making online frame-by-frame analysis challenging. This is especially true for large field-of-view (FOV) imaging. Here, we present CITE-On (Cell Identification and Trace Extraction Online), a convolutional neural network-based algorithm for fast automatic cell identification, segmentation, identity tracking, and trace extraction in two-photon calcium imaging data. CITE-On processes thousands of cells online, including during mesoscopic two-photon imaging, and extracts functional measurements from most neurons in the FOV. Applied to publicly available datasets, the offline version of CITE-On achieves performance similar to that of state-of-the-art methods for offline analysis. Moreover, CITE-On generalizes across calcium indicators, brain regions, and acquisition parameters in anesthetized and awake head-fixed mice. CITE-On represents a powerful tool to speed up image analysis and facilitate closed-loop approaches, for example in combined all-optical imaging and manipulation experiments.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Fast and robust active neuron segmentation in two-photon calcium imaging using spatiotemporal deep learning.Proc Natl Acad Sci U S A. 2019 Apr 23;116(17):8554-8563. doi: 10.1073/pnas.1812995116. Epub 2019 Apr 11. Proc Natl Acad Sci U S A. 2019. PMID: 30975747 Free PMC article.
-
ASTRA: a deep learning algorithm for fast semantic segmentation of large-scale astrocytic networks.bioRxiv [Preprint]. 2023 May 3:2023.05.03.539211. doi: 10.1101/2023.05.03.539211. bioRxiv. 2023. PMID: 37205519 Free PMC article. Preprint.
-
Accurate neuron segmentation method for one-photon calcium imaging videos combining convolutional neural networks and clustering.Commun Biol. 2024 Aug 9;7(1):970. doi: 10.1038/s42003-024-06668-7. Commun Biol. 2024. PMID: 39122882 Free PMC article.
-
Review on Deep Learning Methodologies in Medical Image Restoration and Segmentation.Curr Med Imaging. 2023;19(8):844-854. doi: 10.2174/1573405618666220407112825. Curr Med Imaging. 2023. PMID: 35392788 Review.
-
Learning image-based spatial transformations via convolutional neural networks: A review.Magn Reson Imaging. 2019 Dec;64:142-153. doi: 10.1016/j.mri.2019.05.037. Epub 2019 Jun 11. Magn Reson Imaging. 2019. PMID: 31200026 Review.
Cited by
-
NeuroART: Real-Time Analysis and Targeting of Neuronal Population Activity during Calcium Imaging for Informed Closed-Loop Experiments.eNeuro. 2024 Oct 16;11(10):ENEURO.0079-24.2024. doi: 10.1523/ENEURO.0079-24.2024. Print 2024 Oct. eNeuro. 2024. PMID: 39266327 Free PMC article.
-
Aberration correction in long GRIN lens-based microendoscopes for extended field-of-view two-photon imaging in deep brain regions.Elife. 2025 May 2;13:RP101420. doi: 10.7554/eLife.101420. Elife. 2025. PMID: 40314426 Free PMC article.
-
Pupil engineering for extended depth-of-field imaging in a fluorescence miniscope.Neurophotonics. 2023 Oct;10(4):044302. doi: 10.1117/1.NPh.10.4.044302. Epub 2023 May 8. Neurophotonics. 2023. PMID: 37215637 Free PMC article.
-
CalTrig: A GUI-based Machine Learning Approach for Decoding Neuronal Calcium Transients in Freely Moving Rodents.bioRxiv [Preprint]. 2024 Nov 19:2024.09.30.615860. doi: 10.1101/2024.09.30.615860. bioRxiv. 2024. Update in: eNeuro. 2025 Jul 15;12(7):ENEURO.0009-25.2025. doi: 10.1523/ENEURO.0009-25.2025. PMID: 39372793 Free PMC article. Updated. Preprint.
-
A generative benchmark for evaluating the performance of fluorescent cell image segmentation.Synth Syst Biotechnol. 2024 May 17;9(4):627-637. doi: 10.1016/j.synbio.2024.05.005. eCollection 2024 Dec. Synth Syst Biotechnol. 2024. PMID: 38798889 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

