Interneuron function and cognitive behavior are preserved upon postnatal removal of Lhx6
- PMID: 35318414
- PMCID: PMC8941127
- DOI: 10.1038/s41598-022-09003-4
Interneuron function and cognitive behavior are preserved upon postnatal removal of Lhx6
Abstract
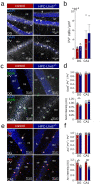
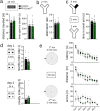
LIM homeobox domain transcription factor 6 (Lhx6) is crucial for the prenatal specification and differentiation of hippocampal GABAergic interneuron precursors. Interestingly, Lhx6 remains to be expressed in parvalbumin-positive hippocampal interneurons (PVIs) long after specification and differentiation have been completed, the functional implications of which remain elusive. We addressed the role of adult-expressed Lhx6 in the hippocampus by knocking down Lhx6 in adult mice (> 8 weeks old) using viral or transgenic expression of Cre-recombinase in Lhx6loxP/loxP mice. Late removal of Lhx6 did not affect the number of PVIs and had no impact on the morphological and physiological properties of PVIs. Furthermore, mice lacking Lhx6 in PVIs displayed normal cognitive behavior. Loss of Lhx6 only partially reduced the expression of Sox6 and Arx, downstream transcription factors that depend on Lhx6 during embryonic development of PVIs. Our data thus suggest that while Lhx6 is vitally important to drive interneuron transcriptional networks during early development, it becomes uncoupled from downstream effectors during postnatal life.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Lhx6 directly regulates Arx and CXCR7 to determine cortical interneuron fate and laminar position.Neuron. 2014 Apr 16;82(2):350-64. doi: 10.1016/j.neuron.2014.02.030. Neuron. 2014. PMID: 24742460 Free PMC article.
-
Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants.J Comp Neurol. 2008 Sep 1;510(1):79-99. doi: 10.1002/cne.21772. J Comp Neurol. 2008. PMID: 18613121 Free PMC article.
-
Knockdown of Lhx6 during embryonic development results in neurophysiological alterations and behavioral deficits analogous to schizophrenia in adult rats.Schizophr Res. 2024 May;267:113-121. doi: 10.1016/j.schres.2024.03.032. Epub 2024 Mar 25. Schizophr Res. 2024. PMID: 38531158 Free PMC article.
-
The development of MGE-derived cortical interneurons: An Lhx6 tale.Int J Dev Biol. 2022;66(1-2-3):43-49. doi: 10.1387/ijdb.210185md. Int J Dev Biol. 2022. PMID: 34881792 Review.
-
Lhx6 and Lhx8: cell fate regulators and beyond.FASEB J. 2015 Oct;29(10):4083-91. doi: 10.1096/fj.14-267500. Epub 2015 Jul 6. FASEB J. 2015. PMID: 26148970 Free PMC article. Review.
Cited by
-
16p11.2 deletion accelerates subpallial maturation and increases variability in human iPSC-derived ventral telencephalic organoids.Development. 2023 Feb 15;150(4):dev201227. doi: 10.1242/dev.201227. Epub 2023 Feb 24. Development. 2023. PMID: 36826401 Free PMC article.
-
Pharmacogenomics of steroid-induced ocular hypertension: relationship to high-tension glaucomas and new pathophysiologic insight.medRxiv [Preprint]. 2025 Aug 13:2025.08.11.25333245. doi: 10.1101/2025.08.11.25333245. medRxiv. 2025. PMID: 40832404 Free PMC article. Preprint.
-
D-serine availability modulates prefrontal cortex inhibitory interneuron development and circuit maturation.Sci Rep. 2023 Jun 13;13(1):9595. doi: 10.1038/s41598-023-35615-5. Sci Rep. 2023. PMID: 37311798 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

