3,4-Dicaffeoylquinic Acid from the Medicinal Plant Ilex kaushue Disrupts the Interaction Between the Five-Fold Axis of Enterovirus A-71 and the Heparan Sulfate Receptor
- PMID: 35319229
- PMCID: PMC9006908
- DOI: 10.1128/jvi.00542-21
3,4-Dicaffeoylquinic Acid from the Medicinal Plant Ilex kaushue Disrupts the Interaction Between the Five-Fold Axis of Enterovirus A-71 and the Heparan Sulfate Receptor
Erratum in
-
Erratum for Hsieh et al., "3,4-Dicaffeoylquinic Acid from the Medicinal Plant Ilex kaushue Disrupts the Interaction between the Five-Fold Axis of Enterovirus A-71 and the Heparan Sulfate Receptor".J Virol. 2022 Jun 8;96(11):e0064922. doi: 10.1128/jvi.00649-22. Epub 2022 May 16. J Virol. 2022. PMID: 35575482 Free PMC article. No abstract available.
Abstract
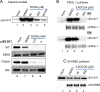
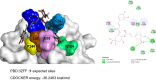
While infections by enterovirus A71 (EV-A71) are generally self-limiting, they can occasionally lead to serious neurological complications and death. No licensed therapies against EV-A71 currently exist. Using anti-virus-induced cytopathic effect assays, 3,4-dicaffeoylquinic acid (3,4-DCQA) from Ilex kaushue extracts was found to exert significant anti-EV-A71 activity, with a broad inhibitory spectrum against different EV-A71 genotypes. Time-of-drug-addition assays revealed that 3,4-DCQA affects the initial phase (entry step) of EV-A71 infection by directly targeting viral particles and disrupting viral attachment to host cells. Using resistant virus selection experiments, we found that 3,4-DCQA targets the glutamic acid residue at position 98 (E98) and the proline residue at position 246 (P246) in the 5-fold axis located within the VP1 structural protein. Recombinant viruses harboring the two mutations were resistant to 3,4-DCQA-elicited inhibition of virus attachment and penetration into human rhabdomyosarcoma (RD) cells. Finally, we showed that 3,4-DCQA specifically inhibited the attachment of EV-A71 to the host receptor heparan sulfate (HS), but not to the scavenger receptor class B member 2 (SCARB2) and P-selectin glycoprotein ligand-1 (PSGL1). Molecular docking analysis confirmed that 3,4-DCQA targets the 5-fold axis to form a stable structure with the E98 and P246 residues through noncovalent and van der Waals interactions. The targeting of E98 and P246 by 3,4-DCQA was found to be specific; accordingly, HS binding of viruses carrying the K242A or K244A mutations in the 5-fold axis was successfully inhibited by 3,4-DCQA.The clinical utility of 3,4-DCQA in the prevention or treatment of EV-A71 infections warrants further scrutiny. IMPORTANCE The canyon region and the 5-fold axis of the EV-A71 viral particle located within the VP1 protein mediate the interaction of the virus with host surface receptors. The three most extensively investigated cellular receptors for EV-A71 include SCARB2, PSGL1, and cell surface heparan sulfate. In the current study, a RD cell-based anti-cytopathic effect assay was used to investigate the potential broad spectrum inhibitory activity of 3,4-DCQA against different EV-A71 strains. Mechanistically, we demonstrate that 3,4-DCQA disrupts the interaction between the 5-fold axis of EV-A71 and its heparan sulfate receptor; however, no effect was seen on the SCARB2 or PSGL1 receptors. Taken together, our findings show that this natural product may pave the way to novel anti-EV-A71 therapeutic strategies.
Keywords: 3; 4-dicaffeoylquinic acid; 5-fold axis; enterovirus-A71; heparan sulphate.
Conflict of interest statement
The authors declare no conflict of interest.
We have no conflicts of interest to declare.
Figures

References
-
- Racaniello VR. 2007. Picornaviridae: the viruses and their replication, p 795–838. In Knipe DavidM, Martin MalcolmA., Martin DianeE, Griffin RobertA, Lamb Bernard, Roizman, Straus Stephen E (ed), Fields virology, 5th ed, vol 1. Lippincott Williams & Wilkins, Philadelphia.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

