Metagenomic Approaches Reveal Strain Profiling and Genotyping of Klebsiella pneumoniae from Hospitalized Patients in China
- PMID: 35319275
- PMCID: PMC9045201
- DOI: 10.1128/spectrum.02190-21
Metagenomic Approaches Reveal Strain Profiling and Genotyping of Klebsiella pneumoniae from Hospitalized Patients in China
Abstract
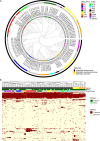
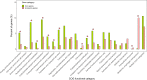
Klebsiella pneumoniae is a leading cause of highly drug-resistant infections in hospitals worldwide. Strain-level bacterial identification on the genetic determinants of multidrug resistance and high pathogenicity is critical for the surveillance and treatment of this clinically relevant pathogen. In this study, metagenomic next-generation sequencing was performed for specimens collected from August 2020 to May 2021 in Ruijin Hospital, Ningbo Women and Children's Hospital, and the Second Affiliated Hospital of Harbin Medical University. Genome biology of K. pneumoniae prevalent in China was characterized based on metagenomic data. Thirty K. pneumoniae strains derived from 14 sequence types were identified by multilocus sequence typing. The hypervirulent ST11 K. pneumoniae strains carrying the KL64 capsular locus were the most prevalent in the hospital population. The phylogenomic analyses revealed that the metagenome-reconstructed strains and public isolate genomes belonging to the same STs were closely related in the phylogenetic tree. Furthermore, the pangenome structure of the detected K. pneumoniae strains was analyzed, particularly focusing on the distribution of antimicrobial resistance genes and virulence genes across the strains. The genes encoding carbapenemases and extended-spectrum beta-lactamases were frequently detected in the strains of ST11 and ST15. The highest numbers of virulence genes were identified in the well-known hypervirulent strains affiliated to ST23 bearing the K1 capsule. In comparison to traditional cultivation and identification, strain-level metagenomics is advantageous to understand the mechanisms underlying resistance and virulence of K. pneumoniae directly from clinical specimens. Our findings should provide novel clues for future research into culture-independent metagenomic surveillance for bacterial pathogens. IMPORTANCE Routine culture and PCR-based molecular testing in the clinical microbiology laboratory are unable to recognize pathogens at the strain level and to detect strain-specific genetic determinants involved in virulence and resistance. To address this issue, we explored the strain-level profiling of K. pneumoniae prevalent in China based on metagenome-sequenced patient materials. Genome biology of the targeted bacterium can be well characterized through decoding sequence signatures and functional gene profiles at the single-strain resolution. The in-depth metagenomic analysis on strain profiling presented here shall provide a promising perspective for culture-free pathogen surveillance and molecular epidemiology of nosocomial infections.
Keywords: K. pneumoniae; MLST; antimicrobial resistance genes; capsule typing; metagenome-reconstructed strains; phylogeny; strain profiling; virulence genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Comparative genomic analysis of the core virulence factor in Klebsiella pneumoniae ST11, based on a global genomic database.Microb Pathog. 2025 Jul;204:107575. doi: 10.1016/j.micpath.2025.107575. Epub 2025 Apr 11. Microb Pathog. 2025. PMID: 40222569
-
High Prevalence of Plasmid-Mediated Quinolone Resistance Determinants Among Serotype K1 Hypervirulent Klebsiella pneumoniae Isolates in China.Microb Drug Resist. 2019 Jun;25(5):681-689. doi: 10.1089/mdr.2018.0173. Epub 2019 Jan 7. Microb Drug Resist. 2019. PMID: 30615560
-
Outbreak of KPC-producing Klebsiella pneumoniae ST15 strains in a Chinese tertiary hospital: resistance and virulence analyses.J Med Microbiol. 2022 Feb;71(2). doi: 10.1099/jmm.0.001494. J Med Microbiol. 2022. PMID: 35156610
-
Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms.Front Cell Infect Microbiol. 2017 Nov 21;7:483. doi: 10.3389/fcimb.2017.00483. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29209595 Free PMC article. Review.
-
Decoding virulence and resistance in Klebsiella pneumoniae: Pharmacological insights, immunological dynamics, and in silico therapeutic strategies.Microb Pathog. 2025 Aug;205:107691. doi: 10.1016/j.micpath.2025.107691. Epub 2025 May 10. Microb Pathog. 2025. PMID: 40355055 Review.
Cited by
-
Genetic characterization of human adenoviruses in patients using metagenomic next-generation sequencing in Hubei, China, from 2018 to 2019.Front Microbiol. 2023 Mar 16;14:1153728. doi: 10.3389/fmicb.2023.1153728. eCollection 2023. Front Microbiol. 2023. PMID: 37007506 Free PMC article.
-
Successful treatment of acute respiratory distress syndrome caused by hypervirulent Klebsiella pneumoniae with extracorporeal membrane oxygenation and continuous renal replacement therapy: A case report and literature review.Front Med (Lausanne). 2022 Aug 24;9:936927. doi: 10.3389/fmed.2022.936927. eCollection 2022. Front Med (Lausanne). 2022. PMID: 36091705 Free PMC article.
-
Hypervirulent Klebsiella pneumoniae (hvKp): Overview, Epidemiology, and Laboratory Detection.Pathog Immun. 2025 Jan 24;10(1):80-119. doi: 10.20411/pai.v10i1.777. eCollection 2024. Pathog Immun. 2025. PMID: 39911145 Free PMC article. Review.
References
-
- Adeolu M, Alnajar S, Naushad S, Gupta RS. 2016. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol 66:5575–5599. doi:10.1099/ijsem.0.001485. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

