Multiple Molecular Dynamics Simulations and Energy Analysis Unravel the Dynamic Properties and Binding Mechanism of Mutants HIV-1 Protease with DRV and CA-p2
- PMID: 35319278
- PMCID: PMC9045218
- DOI: 10.1128/spectrum.00748-21
Multiple Molecular Dynamics Simulations and Energy Analysis Unravel the Dynamic Properties and Binding Mechanism of Mutants HIV-1 Protease with DRV and CA-p2
Abstract
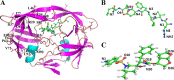
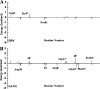
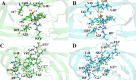
PRS17, a variant of human immunodeficiency virus type I protease (HIV-1 PR), has 17 mutated residues showing high levels of multidrug resistance. To describe the effects of these mutated residues on the dynamic properties and the binding mechanism of PR with substrate and inhibitor, focused on six systems (two complexes of WT PR and PRS17 with inhibitor Darunavir (DRV), two complexes of WT PR and PRS17 with substrate analogue CA-p2, two unligand WT PR and PRS17), we performed multiple molecular dynamics (MD) simulations combined with MM-PBSA and solvated interaction energy (SIE) methods. For both the unligand PRs and ligand-PR complexes, the results from simulations revealed 17 mutated residues alter the flap-flap distance, the distance from flap regions to catalytic sites, and the curling degree of the flap tips. These mutated residues changed the flexibility of the flap region in PR, and thus affected its binding energy with DRV and CA-p2, resulting in differences in sensitivity. Hydrophobic cavity makes an important contribution to the binding of PR and ligands. And most noticeable of all, the binding of the guanidine group in CA-p2 and Arg8' of PRS17 is useful for increasing their binding ability. These results have important guidance for the further design of drugs against multidrug resistant PR. IMPORTANCE Developing effective anti-HIV inhibitors is the current requirement to cope with the emergence of the resistance of mutants. Compared with the experiments, MD simulations along with energy calculations help reduce the time and cost of designing new inhibitors. Based on our simulation results, we propose two factors that may help design effective inhibitors against HIV-1 PR: (i) importance of hydrophobic cavity, and (ii) introduction of polar groups similar to the guanidine group.
Keywords: HIV-1 PR; MD simulation; MM-PBSA analyses; drug resistance; solvated interaction energy analyses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Multiple Molecular Dynamics Simulations and Free-Energy Predictions Uncover the Susceptibility of Variants of HIV-1 Protease against Inhibitors Darunavir and KNI-1657.Langmuir. 2021 Dec 14;37(49):14407-14418. doi: 10.1021/acs.langmuir.1c02348. Epub 2021 Dec 1. Langmuir. 2021. PMID: 34851643
-
Structural and binding insights into HIV-1 protease and P2-ligand interactions through molecular dynamics simulations, binding free energy and principal component analysis.J Mol Graph Model. 2019 Nov;92:112-122. doi: 10.1016/j.jmgm.2019.07.008. Epub 2019 Jul 18. J Mol Graph Model. 2019. PMID: 31351319 Free PMC article.
-
A contribution to the drug resistance mechanism of darunavir, amprenavir, indinavir, and saquinavir complexes with HIV-1 protease due to flap mutation I50V: a systematic MM-PBSA and thermodynamic integration study.J Chem Inf Model. 2013 Aug 26;53(8):2141-53. doi: 10.1021/ci4002102. Epub 2013 Jul 24. J Chem Inf Model. 2013. PMID: 23834142
-
Novel HIV PR inhibitors with C4-substituted bis-THF and bis-fluoro-benzyl target the two active site mutations of highly drug resistant mutant PRS17.Biochem Biophys Res Commun. 2021 Aug 20;566:30-35. doi: 10.1016/j.bbrc.2021.05.094. Epub 2021 Jun 7. Biochem Biophys Res Commun. 2021. PMID: 34111669 Free PMC article.
-
Limiting assumptions in structure-based design: binding entropy.J Comput Aided Mol Des. 2012 Jan;26(1):3-8. doi: 10.1007/s10822-011-9494-1. Epub 2012 Jan 3. J Comput Aided Mol Des. 2012. PMID: 22212342 Free PMC article. Review.
Cited by
-
Insight into the Binding Interaction between PEDCs and hERRγ Utilizing Molecular Docking and Molecular Dynamics Simulations.Molecules. 2024 Jul 10;29(14):3256. doi: 10.3390/molecules29143256. Molecules. 2024. PMID: 39064835 Free PMC article.
References
-
- Henes M, Kosovrasti K, Lockbaum GJ, Leidner F, Nachum GS, Nalivaika EA, Bolon DN, Yilmaz NK, Schiffer CA, Whitfield TW. 2019. Molecular determinants of epistasis in HIV-1 protease: elucidating the interdependence of L89V and L90M mutations in resistance. Biochemistry 58:3711–3726. doi:10.1021/acs.biochem.9b00446. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

