NR1D1 downregulation in astrocytes induces a phenotype that is detrimental to cocultured motor neurons
- PMID: 35319791
- PMCID: PMC9223394
- DOI: 10.1096/fj.202101275R
NR1D1 downregulation in astrocytes induces a phenotype that is detrimental to cocultured motor neurons
Abstract
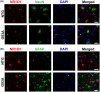
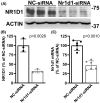
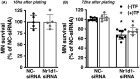
Nuclear receptor subfamily 1 group D member 1 (NR1D1, also known as Rev-erbα) is a nuclear transcription factor that is part of the molecular clock encoding circadian rhythms and may link daily rhythms with metabolism and inflammation. NR1D1, unlike most nuclear receptors, lacks a ligand-dependent activation function domain 2 and is a constitutive transcriptional repressor. Amyotrophic lateral sclerosis (ALS) is the most common adult-onset motor neuron disease, caused by the progressive degeneration of motor neurons in the spinal cord, brain stem, and motor cortex. Approximately 10%-20% of familial ALS is caused by a toxic gain-of-function induced by mutations of the Cu/Zn superoxide dismutase (SOD1). Dysregulated clock and clock-controlled gene expression occur in multiple tissues from mutant hSOD1-linked ALS mouse models. Here we explore NR1D1 dysregulation in the spinal cord of ALS mouse models and its consequences on astrocyte-motor neuron interaction. NR1D1 protein and mRNA expression are significantly downregulated in the spinal cord of symptomatic mice expressing mutant hSOD1, while no changes were observed in age-matched animals overexpressing wild-type hSOD1. In addition, NR1D1 downregulation in primary astrocyte cultures induces a pro-inflammatory phenotype and decreases the survival of cocultured motor neurons. NR1D1 orchestrates the cross talk between physiological pathways identified to be disrupted in ALS (e.g., metabolism, inflammation, redox homeostasis, and circadian rhythms) and we observed that downregulation of NR1D1 alters astrocyte-motor neuron interaction. Our results suggest that NR1D1 could be a potential therapeutic target to prevent astrocyte-mediated motor neuron toxicity in ALS.
Keywords: NR1D1; Rev-erbα; amyotrophic lateral sclerosis; astrocytes; inflammation; motor neurons.
© 2022 The Authors. The FASEB Journal published by Wiley Periodicals LLC on behalf of Federation of American Societies for Experimental Biology.
Figures
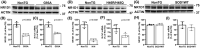


Similar articles
-
Enhancing NAD+ Salvage Pathway Reverts the Toxicity of Primary Astrocytes Expressing Amyotrophic Lateral Sclerosis-linked Mutant Superoxide Dismutase 1 (SOD1).J Biol Chem. 2016 May 13;291(20):10836-46. doi: 10.1074/jbc.M115.698779. Epub 2016 Mar 21. J Biol Chem. 2016. PMID: 27002158 Free PMC article.
-
FABP7 upregulation induces a neurotoxic phenotype in astrocytes.Glia. 2020 Dec;68(12):2693-2704. doi: 10.1002/glia.23879. Epub 2020 Jul 3. Glia. 2020. PMID: 32619303 Free PMC article.
-
Nuclear localization of human SOD1 and mutant SOD1-specific disruption of survival motor neuron protein complex in transgenic amyotrophic lateral sclerosis mice.J Neuropathol Exp Neurol. 2012 Feb;71(2):162-77. doi: 10.1097/NEN.0b013e318244b635. J Neuropathol Exp Neurol. 2012. PMID: 22249462 Free PMC article.
-
Complexity of astrocyte-motor neuron interactions in amyotrophic lateral sclerosis.Neurodegener Dis. 2005;2(3-4):139-46. doi: 10.1159/000089619. Neurodegener Dis. 2005. PMID: 16909019 Review.
-
Rodent Models of Amyotrophic Lateral Sclerosis.Curr Protoc Pharmacol. 2015 Jun 1;69:5.67.1-5.67.21. doi: 10.1002/0471141755.ph0567s69. Curr Protoc Pharmacol. 2015. PMID: 26344214 Free PMC article. Review.
Cited by
-
The role of circadian clock in astrocytes: From cellular functions to ischemic stroke therapeutic targets.Front Neurosci. 2022 Dec 8;16:1013027. doi: 10.3389/fnins.2022.1013027. eCollection 2022. Front Neurosci. 2022. PMID: 36570843 Free PMC article. Review.
-
FABP7 Expression Modulates the Response of Astrocytes to Induced Endotoxemia.Glia. 2025 Aug;73(8):1627-1641. doi: 10.1002/glia.70023. Epub 2025 Apr 18. Glia. 2025. PMID: 40251832 Free PMC article.
References
-
- Yin L, Wu N, Curtin JC, et al. Rev‐erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786‐1789. - PubMed
-
- Renaud JP, Harris JM, Downes M, Burke LJ, Muscat GE. Structure‐function analysis of the Rev‐erbA and RVR ligand‐binding domains reveals a large hydrophobic surface that mediates corepressor binding and a ligand cavity occupied by side chains. Mol Endocrinol. 2000;14:700‐717. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

