A transcriptomics-guided drug target discovery strategy identifies receptor ligands for lung regeneration
- PMID: 35319981
- PMCID: PMC8942365
- DOI: 10.1126/sciadv.abj9949
A transcriptomics-guided drug target discovery strategy identifies receptor ligands for lung regeneration
Abstract
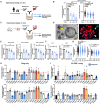
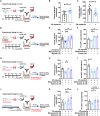
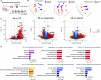
Currently, there is no pharmacological treatment targeting defective tissue repair in chronic disease. Here, we used a transcriptomics-guided drug target discovery strategy using gene signatures of smoking-associated chronic obstructive pulmonary disease (COPD) and from mice chronically exposed to cigarette smoke, identifying druggable targets expressed in alveolar epithelial progenitors, of which we screened the function in lung organoids. We found several drug targets with regenerative potential, of which EP and IP prostanoid receptor ligands had the most profound therapeutic potential in restoring cigarette smoke-induced defects in alveolar epithelial progenitors in vitro and in vivo. Mechanistically, we found, using single-cell RNA sequencing analysis, that circadian clock and cell cycle/apoptosis signaling pathways were differentially expressed in alveolar epithelial progenitor cells in patients with COPD and in a relevant model of COPD, which was prevented by prostaglandin E2 or prostacyclin mimetics. We conclude that specific targeting of EP and IP receptors offers therapeutic potential for injury to repair in COPD.
Figures



References
-
- Basil M. C., Katzen J., Engler A. E., Guo M., Herriges M. J., Kathiriya J. J., Windmueller R., Ysasi A. B., Zacharias W. J., Chapman H. A., Kotton D. N., Rock J. R., Snoeck H. W., Vunjak-Novakovic G., Whitsett J. A., Morrisey E. E., The cellular and physiological basis for lung repair and regeneration: Past, present, and future. Cell Stem Cell 26, 482–502 (2020). - PMC - PubMed
-
- Volckaert T., Yuan T., Chao C. M., Bell H., Sitaula A., Szimmtenings L., El Agha E., Chanda D., Majka S., Bellusci S., Thannickal V. J., Fässler R., De Langhe S. P., Fgf10-hippo epithelial-mesenchymal crosstalk maintains and recruits lung basal stem cells. Dev. Cell 43, 48–59.e5 (2017). - PMC - PubMed
-
- Conlon T. M., John-Schuster G., Heide D., Pfister D., Lehmann M., Hu Y., Ertüz Z., Lopez M. A., Ansari M., Strunz M., Mayr C., Ciminieri C., Costa R., Kohlhepp M. S., Guillot A., Günes G., Jeridi A., Funk M. C., Beroshvili G., Prokosch S., Hetzer J., Verleden S. E., Alsafadi H., Lindner M., Burgstaller G., Becker L., Irmler M., Dudek M., Janzen J., Goffin E., Gosens R., Knolle P., Pirotte B., Stoeger T., Beckers J., Wagner D., Singh I., Theis F. J., de Angelis M. H., O’Connor T., Tacke F., Boutros M., Dejardin E., Eickelberg O., Schiller H. B., Königshoff M., Heikenwalder M., Yildirim A. Ö., Inhibition of LTβR signalling activates WNT-induced regeneration in lung. Nature 588, 151–156 (2020). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

