Sympatric speciation of the spiny mouse from Evolution Canyon in Israel substantiated genomically and methylomically
- PMID: 35320043
- PMCID: PMC9060526
- DOI: 10.1073/pnas.2121822119
Sympatric speciation of the spiny mouse from Evolution Canyon in Israel substantiated genomically and methylomically
Abstract
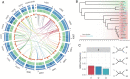
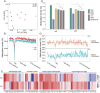
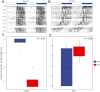
SignificanceWhether sympatric speciation (SS) is rare or common is still debated. Two populations of the spiny mouse, Acomys cahirinus, from Evolution Canyon I (EC I) in Israel have been depicted earlier as speciating sympatrically by molecular markers and transcriptome. Here, we investigated SS both genomically and methylomically, demonstrating that the opposite populations of spiny mice are sister taxa and split from the common ancestor around 20,000 years ago without an allopatric history. Mate choice, olfactory receptors, and speciation genes contributed to prezygotic/postzygotic reproductive isolation. The two populations showed different methylation patterns, facilitating adaptation to their local environment. They cope with abiotic and biotic stresses, due to high solar interslope radiation differences. We conclude that our new genomic and methylomic data substantiated SS.
Keywords: adaptation; genome divergence; methylation; population genetics; sympatric speciation.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Barluenga M., Stölting K. N., Salzburger W., Muschick M., Meyer A., Sympatric speciation in Nicaraguan crater lake cichlid fish. Nature 439, 719–723 (2006). - PubMed
-
- Kondrashov A. S., Kondrashov F. A., Interactions among quantitative traits in the course of sympatric speciation. Nature 400, 351–354 (1999). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

