Modelling aptamers with nucleic acid mimics (NAM): From sequence to three-dimensional docking
- PMID: 35320268
- PMCID: PMC8942228
- DOI: 10.1371/journal.pone.0264701
Modelling aptamers with nucleic acid mimics (NAM): From sequence to three-dimensional docking
Abstract
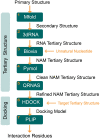
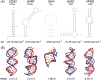
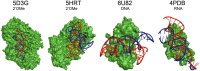
Aptamers are single-stranded oligonucleotides, formerly evolved by Systematic Evolution of Ligands by EXponential enrichment (SELEX), that fold into functional three-dimensional structures. Such conformation is crucial for aptamers' ability to bind to a target with high affinity and specificity. Unnatural nucleotides have been used to develop nucleic acid mimic (NAM) aptamers with increased performance, such as biological stability. Prior knowledge of aptamer-target interactions is critical for applying post-SELEX modifications with unnatural nucleotides since it can affect aptamers' structure and performance. Here, we describe an easy-to-apply in silico workflow using free available software / web servers to predict the tertiary conformation of NAM, DNA and RNA aptamers, as well as the docking with the target molecule. Representative 2'-O-methyl (2'OMe), locked nucleic acid (LNA), DNA and RNA aptamers, with experimental data deposited in Protein Data Bank, were selected to validate the workflow. All aptamers' tertiary structure and docking models were successfully predicted with good structural similarity to the experimental data. Thus, this workflow will boost the development of aptamers, particularly NAM aptamers, by assisting in the rational modification of specific nucleotides and avoiding trial-and-error approaches.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Improving aptamer performance with nucleic acid mimics: de novo and post-SELEX approaches.Trends Biotechnol. 2022 May;40(5):549-563. doi: 10.1016/j.tibtech.2021.09.011. Epub 2021 Oct 28. Trends Biotechnol. 2022. PMID: 34756455 Review.
-
Aptamers: selection, modification and application to nervous system diseases.Curr Med Chem. 2011;18(27):4159-68. doi: 10.2174/092986711797189646. Curr Med Chem. 2011. PMID: 21838689 Review.
-
Artificial Intelligence in Aptamer-Target Binding Prediction.Int J Mol Sci. 2021 Mar 30;22(7):3605. doi: 10.3390/ijms22073605. Int J Mol Sci. 2021. PMID: 33808496 Free PMC article. Review.
-
Post-SELEX optimization of aptamers.Anal Bioanal Chem. 2016 Jul;408(17):4567-73. doi: 10.1007/s00216-016-9556-2. Epub 2016 May 12. Anal Bioanal Chem. 2016. PMID: 27173394 Review.
-
RaptRanker: in silico RNA aptamer selection from HT-SELEX experiment based on local sequence and structure information.Nucleic Acids Res. 2020 Aug 20;48(14):e82. doi: 10.1093/nar/gkaa484. Nucleic Acids Res. 2020. PMID: 32537639 Free PMC article.
Cited by
-
Emerging Approaches for Mitigating Biofilm-Formation-Associated Infections in Farm, Wild, and Companion Animals.Pathogens. 2024 Apr 13;13(4):320. doi: 10.3390/pathogens13040320. Pathogens. 2024. PMID: 38668275 Free PMC article. Review.
-
An overview of structural approaches to study therapeutic RNAs.Front Mol Biosci. 2022 Oct 28;9:1044126. doi: 10.3389/fmolb.2022.1044126. eCollection 2022. Front Mol Biosci. 2022. PMID: 36387283 Free PMC article. Review.
-
Elucidating the molecular docking and binding dynamics of aptamers with spike proteins across SARS-CoV-2 variants of concern.Front Microbiol. 2025 Feb 14;16:1503890. doi: 10.3389/fmicb.2025.1503890. eCollection 2025. Front Microbiol. 2025. PMID: 40028457 Free PMC article.
-
On-site sensing for aflatoxicosis poisoning via ultraviolet excitable aptasensor based on fluorinated ethylene propylene strip: a promising forensic tool.Sci Rep. 2024 Jul 29;14(1):17357. doi: 10.1038/s41598-024-68264-3. Sci Rep. 2024. PMID: 39075202 Free PMC article.
-
Multiplexed In Vivo Screening Using Barcoded Aptamer Technology to Identify Oligonucleotide-Based Targeting Reagents.Nucleic Acid Ther. 2024;34(3):109-124. doi: 10.1089/nat.2024.0010. Epub 2024 May 16. Nucleic Acid Ther. 2024. PMID: 38752363 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

