Population balance modelling captures host cell protein dynamics in CHO cell cultures
- PMID: 35320326
- PMCID: PMC8959726
- DOI: 10.1371/journal.pone.0265886
Population balance modelling captures host cell protein dynamics in CHO cell cultures
Abstract
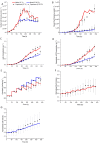
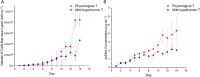
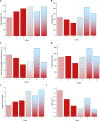
Monoclonal antibodies (mAbs) have been extensively studied for their wide therapeutic and research applications. Increases in mAb titre has been achieved mainly by cell culture media/feed improvement and cell line engineering to increase cell density and specific mAb productivity. However, this improvement has shifted the bottleneck to downstream purification steps. The higher accumulation of the main cell-derived impurities, host cell proteins (HCPs), in the supernatant can negatively affect product integrity and immunogenicity in addition to increasing the cost of capture and polishing steps. Mathematical modelling of bioprocess dynamics is a valuable tool to improve industrial production at fast rate and low cost. Herein, a single stage volume-based population balance model (PBM) has been built to capture Chinese hamster ovary (CHO) cell behaviour in fed-batch bioreactors. Using cell volume as the internal variable, the model captures the dynamics of mAb and HCP accumulation extracellularly under physiological and mild hypothermic culture conditions. Model-based analysis and orthogonal measurements of lactate dehydrogenase activity and double-stranded DNA concentration in the supernatant show that a significant proportion of HCPs found in the extracellular matrix is secreted by viable cells. The PBM then served as a platform for generating operating strategies that optimise antibody titre and increase cost-efficiency while minimising impurity levels.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Mahmuda A, Bande F, Al-Zihiry KJK, Abdulhaleem N, Abd Majid R, Hamat RA, et al. Monoclonal antibodies: A review of therapeutic applications and future prospects. Tropical Journal of Pharmaceutical Research. 2017;16(3):713–22. doi: 10.4314/tjpr.v16i3.29 WOS:000399247000029. - DOI
-
- Xing ZZ, Kenty B, Koyrakh I, Borys M, Pan SH, Li ZJ. Optimizing amino acid composition of CHO cell culture media for a fusion protein production. Process Biochemistry. 2011;46(7):1423–9. doi: 10.1016/j.procbio.2011.03.014 WOS:000292411700006. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

