Charge Engineering Improves the Performance of Bst DNA Polymerase Fusions
- PMID: 35320674
- PMCID: PMC9514385
- DOI: 10.1021/acssynbio.1c00559
Charge Engineering Improves the Performance of Bst DNA Polymerase Fusions
Abstract
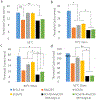
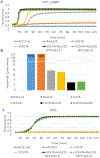
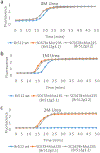
The charge states of proteins can greatly influence their stabilities and interactions with substrates, and the addition of multiple charges (supercharging) has been shown to be a successful approach for engineering protein stability and function. The addition of a fast-folding fusion domain to the Bacillus stearothermophilus DNA polymerase improved its functionality in isothermal amplification assays, and further charge engineering of this domain has increased both protein stability and diagnostics performance. When combined with mutations that stabilize the core of the protein, the charge-engineered fusion domain leads to the ability to carry out loop-mediated isothermal amplification (LAMP) at temperatures up to 74° C or in the presence of high concentrations of urea, with detection times under 10 min. Adding both positive and negative charges to the fusion domain led to changes in the relative reverse transcriptase and DNA polymerase activities of the polymerase. Overall, the development of a modular fusion domain whose charged surface can be modified at will should prove to be of use in the engineering of other polymerases and, in general, may prove useful for protein stabilization.
Keywords: Br512; charge engineering; fusion domains; high-temperature LAMP; polymerase engineering; supercharging.
Conflict of interest statement
The authors declare the following competing financial interest(s): IP, SB, and AE are named inventors on an IP filing relating to methods and compositions described in this manuscript.
Figures

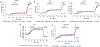
Similar articles
-
Double domain fusion improves the reverse transcriptase activity and inhibitor tolerance of Bst DNA polymerase.Int J Biol Macromol. 2024 Aug;274(Pt 1):133243. doi: 10.1016/j.ijbiomac.2024.133243. Epub 2024 Jun 18. Int J Biol Macromol. 2024. PMID: 38901507
-
Improved Bst DNA Polymerase Variants Derived via a Machine Learning Approach.Biochemistry. 2023 Jan 17;62(2):410-418. doi: 10.1021/acs.biochem.1c00451. Epub 2021 Nov 11. Biochemistry. 2023. PMID: 34762799 Free PMC article.
-
Improvement of φ29 DNA polymerase amplification performance by fusion of DNA binding motifs.Proc Natl Acad Sci U S A. 2010 Sep 21;107(38):16506-11. doi: 10.1073/pnas.1011428107. Epub 2010 Sep 7. Proc Natl Acad Sci U S A. 2010. PMID: 20823261 Free PMC article.
-
Bst polymerase - a humble relative of Taq polymerase.Comput Struct Biotechnol J. 2023 Sep 12;21:4519-4535. doi: 10.1016/j.csbj.2023.09.008. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37767105 Free PMC article. Review.
-
Engineering Polymerases for New Functions.Trends Biotechnol. 2019 Oct;37(10):1091-1103. doi: 10.1016/j.tibtech.2019.03.011. Epub 2019 Apr 16. Trends Biotechnol. 2019. PMID: 31003719 Free PMC article. Review.
Cited by
-
Research progress of loop-mediated isothermal amplification in the detection of Salmonella for food safety applications.Discov Nano. 2024 Aug 6;19(1):124. doi: 10.1186/s11671-024-04075-9. Discov Nano. 2024. PMID: 39105889 Free PMC article. Review.
-
Loop-Mediated Isothermal Amplification: From Theory to Practice.Russ J Bioorg Chem. 2022;48(6):1159-1174. doi: 10.1134/S106816202206022X. Epub 2022 Dec 23. Russ J Bioorg Chem. 2022. PMID: 36590469 Free PMC article.
-
Supercharged fluorescent proteins detect lanthanides via direct antennae signaling.Nat Commun. 2024 Oct 24;15(1):9200. doi: 10.1038/s41467-024-53106-7. Nat Commun. 2024. PMID: 39448572 Free PMC article.
-
Kinetics of elementary steps in loop-mediated isothermal amplification (LAMP) show that strand invasion during initiation is rate-limiting.Nucleic Acids Res. 2023 Jan 11;51(1):488-499. doi: 10.1093/nar/gkac1221. Nucleic Acids Res. 2023. PMID: 36583345 Free PMC article.
-
Strategies and procedures to generate chimeric DNA polymerases for improved applications.Appl Microbiol Biotechnol. 2024 Aug 21;108(1):445. doi: 10.1007/s00253-024-13276-2. Appl Microbiol Biotechnol. 2024. PMID: 39167106 Free PMC article. Review.
References
-
- Strickler SS; Gribenko AV; Keiffer TR; Tomlinson J; Reihle T; Loladze VV; Makhatadze GI; et al. Protein stability and surface electrostatics: a charged relationship. Biochemistry 2006, 45, 2761–2766. - PubMed
-
- Simeonov P; Berger-Hoffmann R; Hoffmann R; Sträter N; Zuchner T Surface supercharged human enteropeptidase light chain shows improved solubility and refolding yield. Protein Eng., Des. Sel 2011, 24, 261–268. - PubMed
-
- Miklos AE; Kluwe C; Der BS; Pai S; Sircar A; Hughes RA; Berrondo M; Xu J; Codrea V; Buckley PE; Calm AM; Welsh HS; Warner CR; Zacharko MA; Carney JP; Gray JJ; Georgiou G; Kuhlman B; Ellington AD Structure-based design of supercharged, highly thermoresistant antibodies. Chem. Biol 2012, 19, 449–455. - PMC - PubMed
-
- Austerberry JI; Thistlethwaite A; Fisher K; Golovanov AP; Pluen A; Esfandiary R; van der Walle CF; Warwicker J; Derrick JP; Curtis R Arginine to Lysine Mutations Increase the Aggregation Stability of a Single-Chain Variable Fragment through Unfolded-State Interactions. Biochemistry 2019, 58, 3413–3421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

