AT-NeuroEAE: A Joint Extraction Model of Events With Attributes for Research Sharing-Oriented Neuroimaging Provenance Construction
- PMID: 35321479
- PMCID: PMC8936590
- DOI: 10.3389/fnins.2021.739535
AT-NeuroEAE: A Joint Extraction Model of Events With Attributes for Research Sharing-Oriented Neuroimaging Provenance Construction
Abstract
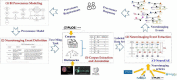
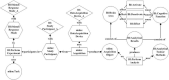
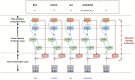
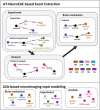
Provenances are a research focus of neuroimaging resources sharing. An amount of work has been done to construct high-quality neuroimaging provenances in a standardized and convenient way. However, besides existing processed-based provenance extraction methods, open research sharing in computational neuroscience still needs one way to extract provenance information from rapidly growing published resources. This paper proposes a literature mining-based approach for research sharing-oriented neuroimaging provenance construction. A group of neuroimaging event-containing attributes are defined to model the whole process of neuroimaging researches, and a joint extraction model based on deep adversarial learning, called AT-NeuroEAE, is proposed to realize the event extraction in a few-shot learning scenario. Finally, a group of experiments were performed on the real data set from the journal PLOS ONE. Experimental results show that the proposed method provides a practical approach to quickly collect research information for neuroimaging provenance construction oriented to open research sharing.
Keywords: attribute extraction; deep adversarial learning; event extraction; neuroimaging provenance; neuroimaging text mining.
Copyright © 2022 Lin, Xu, Sheng, Chen and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Abacha A. B., Herrera A., Ke W., Long L. R., Antani S., Demner-Fushman D. (2017). “Named entity recognition in functional neuroimaging literature,” in Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, 2218–2220. 10.1109/BIBM.2017.8218002 - DOI
-
- Abera B. (2020). Event extraction and representation model from news articles. Int. J. Innov. Eng. Technol. 16 1–8.
-
- Abrams M. B., Bjaalie J. G., Das S., Egan G. F., Martone M. E. (2021). A standards organization for open and fair neuroscience: the international neuroinformatics coordinating facility. Neuroinformatics 1–12. 10.31219/6osf.io/3rt9b - DOI - PMC - PubMed
-
- Agirre A. G., Marimo M., Intxaurrondo A., Rabal O., Krallinger M. (2019). “PharmaCoNER: pharmacological substances, compounds and proteins named entity recognition track,” in Proceedings of the 5th Workshop on BioNLP Open Shared Tasks, Hong Kong.
LinkOut - more resources
Full Text Sources

