Highly Sensitive Lineage Discrimination of SARS-CoV-2 Variants through Allele-Specific Probe PCR
- PMID: 35321556
- PMCID: PMC9020347
- DOI: 10.1128/jcm.02283-21
Highly Sensitive Lineage Discrimination of SARS-CoV-2 Variants through Allele-Specific Probe PCR
Abstract
Tools to detect SARS-CoV-2 variants of concern and track the ongoing evolution of the virus are necessary to support public health efforts and the design and evaluation of novel COVID-19 therapeutics and vaccines. Although next-generation sequencing (NGS) has been adopted as the gold standard method for discriminating SARS-CoV-2 lineages, alternative methods may be required when processing samples with low viral loads or low RNA quality. To this aim, an allele-specific probe PCR (ASP-PCR) targeting lineage-specific single nucleotide polymorphisms (SNPs) was developed and used to screen 1,082 samples from two clinical trials in the United Kingdom and Brazil. Probit regression models were developed to compare ASP-PCR performance against 1,771 NGS results for the same cohorts. Individual SNPs were shown to readily identify specific variants of concern. ASP-PCR was shown to discriminate SARS-CoV-2 lineages with a higher likelihood than NGS over a wide range of viral loads. The comparative advantage for ASP-PCR over NGS was most pronounced in samples with cycle threshold (CT) values between 26 and 30 and in samples that showed evidence of degradation. Results for samples screened by ASP-PCR and NGS showed 99% concordant results. ASP-PCR is well suited to augment but not replace NGS. The method can differentiate SARS-CoV-2 lineages with high accuracy and would be best deployed to screen samples with lower viral loads or that may suffer from degradation. Future work should investigate further destabilization from primer-target base mismatch through altered oligonucleotide chemistry or chemical additives.
Keywords: SARS-CoV-2; allele-specific probe PCR; diagnostics; next-generation sequencing; variant identification; variants of concern.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
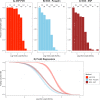


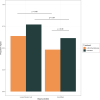
References
-
- World Health Organization. 2021. Genomic sequencing of SARS-CoV-2: a guide to implementation for maximum impact on public health. World Health Organization, Geneva, Switzerland.
-
- Lythgoe KA, Hall M, Ferretti L, de Cesare M, MacIntyre-Cockett G, Trebes A, Andersson M, Otecko N, Wise EL, Moore N, Lynch J, Kidd S, Cortes N, Mori M, Williams R, Vernet G, Justice A, Green A, Nicholls SM, Ansari MA, Abeler-Dörner L, Moore CE, Peto TEA, Eyre DW, Shaw R, Simmonds P, Buck D, Todd JA, on behalf of the Oxford Virus Sequencing Analysis Group (OVSG), Connor TR, Ashraf S, da Silva Filipe A, Shepherd J, Thomson EC, The COVID-19 Genomics UK (COG-UK) Consortium , Bonsall D, Fraser C, Golubchik T. 2021. SARS-CoV-2 within-host diversity and transmission. Science 372:eabg0821. 10.1126/science.abg0821. - DOI - PMC - PubMed
-
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Sheffield COVID-19 Genomics Group, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. 2020. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.e19. 10.1016/j.cell.2020.06.043. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

