Simple and efficient isolation of plant genomic DNA using magnetic ionic liquids
- PMID: 35321738
- PMCID: PMC8943943
- DOI: 10.1186/s13007-022-00860-8
Simple and efficient isolation of plant genomic DNA using magnetic ionic liquids
Abstract
Background: Plant DNA isolation and purification is a time-consuming and laborious process relative to epithelial and viral DNA sample preparation due to the cell wall. The lysis of plant cells to free intracellular DNA normally requires high temperatures, chemical surfactants, and mechanical separation of plant tissue prior to a DNA purification step. Traditional DNA purification methods also do not aid themselves towards fieldwork due to the numerous chemical and bulky equipment requirements.
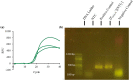
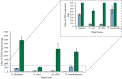
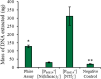
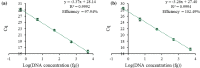
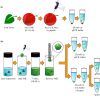
Results: In this study, intact plant tissue was coated by hydrophobic magnetic ionic liquids (MILs) and ionic liquids (ILs) and allowed to incubate under static conditions or dispersed in a suspension buffer to facilitate cell disruption and DNA extraction. The DNA-enriched MIL or IL was successfully integrated into the qPCR buffer without inhibiting the reaction. The two aforementioned advantages of ILs and MILs allow plant DNA sample preparation to occur in one minute or less without the aid of elevated temperatures or chemical surfactants that typically inhibit enzymatic amplification methods. MIL or IL-coated plant tissue could be successfully integrated into a qPCR assay without the need for custom enzymes or manual DNA isolation/purification steps that are required for conventional methods.
Conclusions: The limited amount of equipment, chemicals, and time required to disrupt plant cells while simultaneously extracting DNA using MILs makes the described procedure ideal for fieldwork and lab work in low resource environments.
Keywords: Aloe vera L.; Arabidopsis thaliana (L.) Heynh.; Ionic liquids; Nicotiana benthaminana Domin; One-pot qPCR; One-step cell lysis; Plant DNA isolation; Quercus alba L..
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Meyer R. Development and application of DNA analytical methods for the detection of GMOs in food. Food Control. 1999;10:391–399. doi: 10.1016/S0956-7135(99)00081-X. - DOI
-
- Emaus MN, Varona M, Eitzmann DR, Hsieh SA, Zeger VR, Anderson JL. Nucleic acid extraction: fundamentals of sample preparation methodologies, current advancements, and future endeavors. TrAC - Trends Anal Chem. 2020;130:115985. doi: 10.1016/j.trac.2020.115985. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

