Disruption of GMNC-MCIDAS multiciliogenesis program is critical in choroid plexus carcinoma development
- PMID: 35322202
- PMCID: PMC9345885
- DOI: 10.1038/s41418-022-00950-z
Disruption of GMNC-MCIDAS multiciliogenesis program is critical in choroid plexus carcinoma development
Abstract
Multiciliated cells (MCCs) in the brain reside in the ependyma and the choroid plexus (CP) epithelia. The CP secretes cerebrospinal fluid that circulates within the ventricular system, driven by ependymal cilia movement. Tumors of the CP are rare primary brain neoplasms mostly found in children. CP tumors exist in three forms: CP papilloma (CPP), atypical CPP, and CP carcinoma (CPC). Though CPP and atypical CPP are generally benign and can be resolved by surgery, CPC is a particularly aggressive and little understood cancer with a poor survival rate and a tendency for recurrence and metastasis. In contrast to MCCs in the CP epithelia, CPCs in humans are characterized by solitary cilia, frequent TP53 mutations, and disturbances to multiciliogenesis program directed by the GMNC-MCIDAS transcriptional network. GMNC and MCIDAS are early transcriptional regulators of MCC fate differentiation in diverse tissues. Consistently, components of the GMNC-MCIDAS transcriptional program are expressed during CP development and required for multiciliation in the CP, while CPC driven by deletion of Trp53 and Rb1 in mice exhibits multiciliation defects consequent to deficiencies in the GMNC-MCIDAS program. Previous studies revealed that abnormal NOTCH pathway activation leads to CPP. Here we show that combined defects in NOTCH and Sonic Hedgehog signaling in mice generates tumors that are similar to CPC in humans. NOTCH-driven CP tumors are monociliated, and disruption of the NOTCH complex restores multiciliation and decreases tumor growth. NOTCH suppresses multiciliation in tumor cells by inhibiting the expression of GMNC and MCIDAS, while Gmnc-Mcidas overexpression rescues multiciliation defects and suppresses tumor cell proliferation. Taken together, these findings indicate that reactivation of the GMNC-MCIDAS multiciliogenesis program is critical for inhibiting tumorigenesis in the CP, and it may have therapeutic implications for the treatment of CPC.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




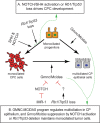
References
-
- Nonami Y, Narita K, Nakamura H, Inoue T, Takeda S. Developmental changes in ciliary motility on choroid plexus epithelial cells during the perinatal period. Cytoskeleton. 2013;70:797–803. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

