Transcriptome-wide identification of RNA-binding protein binding sites using seCLIP-seq
- PMID: 35322209
- PMCID: PMC11134598
- DOI: 10.1038/s41596-022-00680-z
Transcriptome-wide identification of RNA-binding protein binding sites using seCLIP-seq
Abstract
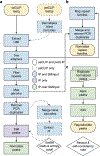
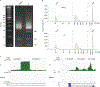
Discovery of interaction sites between RNA-binding proteins (RBPs) and their RNA targets plays a critical role in enabling our understanding of how these RBPs control RNA processing and regulation. Cross-linking and immunoprecipitation (CLIP) provides a generalizable, transcriptome-wide method by which RBP/RNA complexes are purified and sequenced to identify sites of intermolecular contact. By simplifying technical challenges in prior CLIP methods and incorporating the generation of and quantitative comparison against size-matched input controls, the single-end enhanced CLIP (seCLIP) protocol allows for the profiling of these interactions with high resolution, efficiency and scalability. Here, we present a step-by-step guide to the seCLIP method, detailing critical steps and offering insights regarding troubleshooting and expected results while carrying out the ~4-d protocol. Furthermore, we describe a comprehensive bioinformatics pipeline that offers users the tools necessary to process two replicate datasets and identify reproducible and significant peaks for an RBP of interest in ~2 d.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
G.W.Y. is co-founder, member of the Board of Directors, on the Science Advisory Board, an equity holder and a paid consultant for Locanabio and Eclipse BioInnovations. G.W.Y. is a visiting professor at the National University of Singapore. G.W.Y.’s interest(s) have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. A.A.S. is co-founder and Research & Development Director for Eclipse Bioinnovations. E.L.V.N. is co-founder, member of the Board of Directors, on the Science Advisory Board, an equity holder and a paid consultant for Eclipse BioInnovations. E.L.V.N.’s interest(s) have been reviewed and approved by Baylor College of Medicine in accordance with its conflict-of-interest policies. The authors declare no other competing interests.
Figures

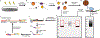


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

