AAV-delivered suppressor tRNA overcomes a nonsense mutation in mice
- PMID: 35322228
- PMCID: PMC9446716
- DOI: 10.1038/s41586-022-04533-3
AAV-delivered suppressor tRNA overcomes a nonsense mutation in mice
Abstract
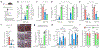
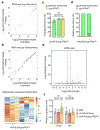
Gene therapy is a potentially curative medicine for many currently untreatable diseases, and recombinant adeno-associated virus (rAAV) is the most successful gene delivery vehicle for in vivo applications1-3. However, rAAV-based gene therapy suffers from several limitations, such as constrained DNA cargo size and toxicities caused by non-physiological expression of a transgene4-6. Here we show that rAAV delivery of a suppressor tRNA (rAAV.sup-tRNA) safely and efficiently rescued a genetic disease in a mouse model carrying a nonsense mutation, and effects lasted for more than 6 months after a single treatment. Mechanistically, this was achieved through a synergistic effect of premature stop codon readthrough and inhibition of nonsense-mediated mRNA decay. rAAV.sup-tRNA had a limited effect on global readthrough at normal stop codons and did not perturb endogenous tRNA homeostasis, as determined by ribosome profiling and tRNA sequencing, respectively. By optimizing the AAV capsid and the route of administration, therapeutic efficacy in various target tissues was achieved, including liver, heart, skeletal muscle and brain. This study demonstrates the feasibility of developing a toolbox of AAV-delivered nonsense suppressor tRNAs operating on premature termination codons (AAV-NoSTOP) to rescue pathogenic nonsense mutations and restore gene function under endogenous regulation. As nonsense mutations account for 11% of pathogenic mutations, AAV-NoSTOP can benefit a large number of patients. AAV-NoSTOP obviates the need to deliver a full-length protein-coding gene that may exceed the rAAV packaging limit, elicit adverse immune responses or cause transgene-related toxicities. It therefore represents a valuable addition to gene therapeutics.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
COMPETING INTERESTS
J.W., Y.Z., C.A.M., G.G. and D.W. are inventors of a patent application filed by University of Massachusetts Chan Medical School concerning the design and applications of suppressor tRNAs described in this study. G.G. is a scientific co-founder of Voyager Therapeutics, Adrenas Therapeutics and Aspa Therapeutics and holds equity in the companies. G.G. and D.W. are inventors on patents related to AAV-based gene therapy, some of which were licensed to commercial entities. Other authors have no competing interest.
Figures
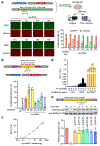
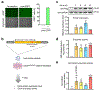
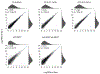
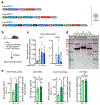
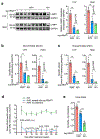
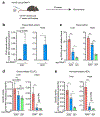
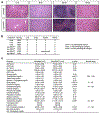



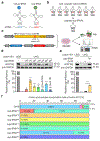

Comment in
-
tRNA molecules run a STOP sign.Nat Rev Drug Discov. 2022 May;21(5):337. doi: 10.1038/d41573-022-00066-6. Nat Rev Drug Discov. 2022. PMID: 35396352 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

