Androgen receptor activity in T cells limits checkpoint blockade efficacy
- PMID: 35322234
- PMCID: PMC10294141
- DOI: 10.1038/s41586-022-04522-6
Androgen receptor activity in T cells limits checkpoint blockade efficacy
Abstract
Immune checkpoint blockade has revolutionized the field of oncology, inducing durable anti-tumour immunity in solid tumours. In patients with advanced prostate cancer, immunotherapy treatments have largely failed1-5. Androgen deprivation therapy is classically administered in these patients to inhibit tumour cell growth, and we postulated that this therapy also affects tumour-associated T cells. Here we demonstrate that androgen receptor (AR) blockade sensitizes tumour-bearing hosts to effective checkpoint blockade by directly enhancing CD8 T cell function. Inhibition of AR activity in CD8 T cells prevented T cell exhaustion and improved responsiveness to PD-1 targeted therapy via increased IFNγ expression. AR bound directly to Ifng and eviction of AR with a small molecule significantly increased cytokine production in CD8 T cells. Together, our findings establish that T cell intrinsic AR activity represses IFNγ expression and represents a novel mechanism of immunotherapy resistance.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
R.F.T. and J.N.G. are employees of the US Government. The contents do not represent the views of the US Department of Veterans Affairs or the United States Government. S.A.H. is an employee of AstraZeneca. A.E.M. received research funding from AstraZeneca.
Figures



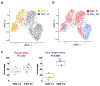



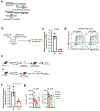

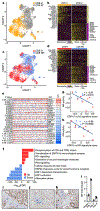


Comment in
-
AR in immunotherapy.Nat Rev Cancer. 2022 Jun;22(6):319. doi: 10.1038/s41568-022-00476-z. Nat Rev Cancer. 2022. PMID: 35393540 No abstract available.
-
Androgen effects cause sex-biased impairment of CD8+ T cell antitumor activity.Sci Immunol. 2022 Jul;7(73):eabq6783. doi: 10.1126/sciimmunol.abq6783. Epub 2022 Jul 1. Sci Immunol. 2022. PMID: 35776801
References
-
- Beer TM et al. Randomized, double-blind, phase III trial of ipilimumab versus placebo in asymptomatic or minimally symptomatic patients with metastatic chemotherapy-naive castration-resistant prostate cancer. J. Clin. Oncol 35, 40–47 (2017). - PubMed
-
- Fong PC et al. Pembrolizumab plus enzalutamide in abiraterone-pretreated patients with metastatic castrate resistant prostate cancer: cohort C of the phase 1b/2 KEYNOTE-365 study. J. Clin. Oncol 37, suppl:abstr 5010 (2019)
-
- Sharma P et al. Nivolumab plus ipilimumab for metastatic castration-resistant prostate cancer: preliminary analysis of patients in the CheckMate 650 trial. Cancer Cell. 38, 489–499 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

