Mechanism of action and therapeutic route for a muscular dystrophy caused by a genetic defect in lipid metabolism
- PMID: 35322809
- PMCID: PMC8943011
- DOI: 10.1038/s41467-022-29270-z
Mechanism of action and therapeutic route for a muscular dystrophy caused by a genetic defect in lipid metabolism
Abstract
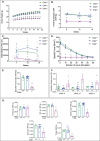
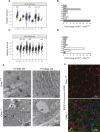
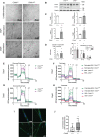
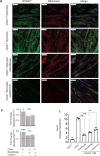
CHKB encodes one of two mammalian choline kinase enzymes that catalyze the first step in the synthesis of the membrane phospholipid phosphatidylcholine. In humans and mice, inactivation of the CHKB gene (Chkb in mice) causes a recessive rostral-to-caudal muscular dystrophy. Using Chkb knockout mice, we reveal that at no stage of the disease is phosphatidylcholine level significantly altered. We observe that in affected muscle a temporal change in lipid metabolism occurs with an initial inability to utilize fatty acids for energy via mitochondrial β-oxidation resulting in shunting of fatty acids into triacyglycerol as the disease progresses. There is a decrease in peroxisome proliferator-activated receptors and target gene expression specific to Chkb-/- affected muscle. Treatment of Chkb-/- myocytes with peroxisome proliferator-activated receptor agonists enables fatty acids to be used for β-oxidation and prevents triacyglyerol accumulation, while simultaneously increasing expression of the compensatory choline kinase alpha (Chka) isoform, preventing muscle cell injury.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Ishidate K. Choline/ethanolamine kinase from mammalian tissues. Biochim Biophys. Acta. 1997;1348:70–78. - PubMed
-
- Aoyama C, Liao H, Ishidate K. Structure and function of choline kinase isoforms in mammalian cells. Prog. Lipid Res. 2004;43:266–281. - PubMed
-
- Bansal D, et al. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 2003;423:168–172. - PubMed
-
- Sher RB, et al. A rostrocaudal muscular dystrophy caused by a defect in choline kinase beta, the first enzyme in phosphatidylcholine biosynthesis. J. Biol. Chem. 2006;281:4938–4948. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

