Proteasomal turnover of the RhoGAP tumor suppressor DLC1 is regulated by HECTD1 and USP7
- PMID: 35322810
- PMCID: PMC8943137
- DOI: 10.1038/s41598-022-08844-3
Proteasomal turnover of the RhoGAP tumor suppressor DLC1 is regulated by HECTD1 and USP7
Abstract
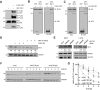
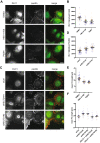
The Rho GTPase activating protein Deleted in Liver Cancer 1 (DLC1) is frequently downregulated through genetic and epigenetic mechanisms in various malignancies, leading to aberrant Rho GTPase signaling and thus facilitating cancer progression. Here we show that in breast cancer cells, dysregulation of DLC1 expression occurs at the protein level through rapid degradation via the ubiquitin-proteasome system. Using mass spectrometry, we identify two novel DLC1 interaction partners, the ubiquitin-ligase HECTD1 and the deubiquitinating enzyme ubiquitin-specific-processing protease 7 (USP7). While DLC1 protein expression was rapidly downregulated upon pharmacological inhibition of USP7, siRNA-mediated knockdown of HECTD1 increased DLC1 protein levels and impaired its degradation. Immunofluorescence microscopy analyses revealed that the modulation of HECTD1 levels and USP7 activity altered DLC1 abundance at focal adhesions, its primary site of action. Thus, we propose opposing regulatory mechanisms of DLC1 protein homeostasis by USP7 and HECTD1, which could open up strategies to counteract downregulation and restore DLC1 expression in cancer.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Inhibition of cytoplasmic EZH2 induces antitumor activity through stabilization of the DLC1 tumor suppressor protein.Nat Commun. 2021 Dec 3;12(1):6941. doi: 10.1038/s41467-021-26993-3. Nat Commun. 2021. PMID: 34862367 Free PMC article.
-
The intracellular stability of DLC1 is regulated by the 26S proteasome in human hepatocellular carcinoma cell line Hep3B.Biochem Biophys Res Commun. 2011 Jan 7;404(1):279-83. doi: 10.1016/j.bbrc.2010.11.107. Epub 2010 Dec 2. Biochem Biophys Res Commun. 2011. PMID: 21130076
-
CRL4A-FBXW5-mediated degradation of DLC1 Rho GTPase-activating protein tumor suppressor promotes non-small cell lung cancer cell growth.Proc Natl Acad Sci U S A. 2013 Oct 15;110(42):16868-73. doi: 10.1073/pnas.1306358110. Epub 2013 Sep 30. Proc Natl Acad Sci U S A. 2013. PMID: 24082123 Free PMC article.
-
Tumor suppressor gene DLC1: Its modifications, interactive molecules, and potential prospects for clinical cancer application.Int J Biol Macromol. 2021 Jul 1;182:264-275. doi: 10.1016/j.ijbiomac.2021.04.022. Epub 2021 Apr 6. Int J Biol Macromol. 2021. PMID: 33836193 Review.
-
Regulation of deleted in liver cancer 1 tumor suppressor by protein-protein interactions and phosphorylation.Int J Cancer. 2014 Jul 15;135(2):264-9. doi: 10.1002/ijc.28505. Epub 2013 Oct 17. Int J Cancer. 2014. PMID: 24114040 Review.
Cited by
-
The pro-oncogenic noncanonical activity of a RAS•GTP:RanGAP1 complex facilitates nuclear protein export.Nat Cancer. 2024 Dec;5(12):1902-1918. doi: 10.1038/s43018-024-00847-5. Epub 2024 Nov 11. Nat Cancer. 2024. PMID: 39528835 Free PMC article.
References
-
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. - PubMed
-
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on Rho GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 2005;6:167–180. - PubMed
-
- Hodge RG, Ridley AJ. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 2016;17:496–510. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

