GENOMES UNCOUPLED1 plays a key role during the de-etiolation process in Arabidopsis
- PMID: 35322876
- PMCID: PMC9324965
- DOI: 10.1111/nph.18115
GENOMES UNCOUPLED1 plays a key role during the de-etiolation process in Arabidopsis
Abstract
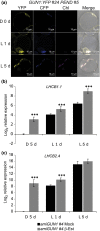
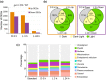
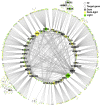
One of the most dramatic challenges in the life of a plant occurs when the seedling emerges from the soil and exposure to light triggers expression of genes required for establishment of photosynthesis. This process needs to be tightly regulated, as premature accumulation of light-harvesting proteins and photoreactive Chl precursors causes oxidative damage when the seedling is first exposed to light. Photosynthesis genes are encoded by both nuclear and plastid genomes, and to establish the required level of control, plastid-to-nucleus (retrograde) signalling is necessary to ensure correct gene expression. We herein show that a negative GENOMES UNCOUPLED1 (GUN1)-mediated retrograde signal restricts chloroplast development in darkness and during early light response by regulating the transcription of several critical transcription factors linked to light response, photomorphogenesis, and chloroplast development, and consequently their downstream target genes in Arabidopsis. Thus, the plastids play an essential role during skotomorphogenesis and the early light response, and GUN1 acts as a safeguard during the critical step of seedling emergence from darkness.
Keywords: GUN1; chloroplast; greening; light signalling; plastid retrograde signalling; transcriptional regulation.
© 2022 The Authors New Phytologist © 2022 New Phytologist Foundation.
Figures





References
-
- Alabadí D, Gallego‐Bartolomé J, Orlando L, García‐Cárcel L, Rubio V, Martínez C, Frigerio M, Iglesias‐Pedraz JM, Espinosa A, Deng XW et al. 2008. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de‐etiolation in darkness. The Plant Journal 53: 324–335. - PubMed
-
- Castillo‐Davis CI, Hartl DL. 2003. GeneMerge – post‐genomic analysis, data mining, and hypothesis testing. Bioinformatics 19: 891–892. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

