In Silico Evaluation of Antifungal Compounds from Marine Sponges against COVID-19-Associated Mucormycosis
- PMID: 35323514
- PMCID: PMC8950821
- DOI: 10.3390/md20030215
In Silico Evaluation of Antifungal Compounds from Marine Sponges against COVID-19-Associated Mucormycosis
Abstract
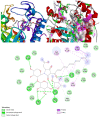
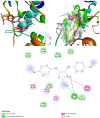
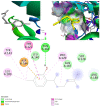
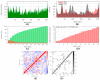
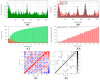
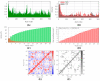
The world is already facing the devastating effects of the SARS-CoV-2 pandemic. A disseminated mucormycosis epidemic emerged to worsen this situation, causing havoc, especially in India. This research aimed to perform a multitargeted docking study of marine-sponge-origin bioactive compounds against mucormycosis. Information on proven drug targets and marine sponge compounds was obtained via a literature search. A total of seven different targets were selected. Thirty-five compounds were chosen using the PASS online program. For homology modeling and molecular docking, FASTA sequences and 3D structures for protein targets were retrieved from NCBI and PDB databases. Autodock Vina in PyRx 0.8 was used for docking studies. Further, molecular dynamics simulations were performed using the IMODS server for top-ranked docked complexes. Moreover, the drug-like properties and toxicity analyses were performed using Lipinski parameters in Swiss-ADME, OSIRIS, ProTox-II, pkCSM, and StopTox servers. The results indicated that naamine D, latrunculin A and S, (+)-curcudiol, (+)-curcuphenol, aurantoside I, and hyrtimomine A had the highest binding affinity values of -8.8, -8.6, -9.8, -11.4, -8.0, -11.4, and -9.0 kcal/mol, respectively. In sum, all MNPs included in this study are good candidates against mucormycosis. (+)-curcudiol and (+)-curcuphenol are promising compounds due to their broad-spectrum target inhibition potential.
Keywords: CAM; COVID-19; SARS-CoV-2; antifungal; antiviral; bioactive compounds; disseminated mucormycosis; marine drugs; marine sponges; molecular docking.
Conflict of interest statement
The authors declare no conflict of interest.
Figures













References
-
- Guzmán-Castro S., Chora-Hernandez L.D., Trujillo-Alonso G., Calvo-Villalobos I., Sanchez-Rangel A., Ferrer-Alpuin E., Ruiz-Jimenez M., Corzo-Leon D.E. COVID-19-associated mucormycosis, diabetes and steroid therapy: Experience in a single centre in Western Mexico. Mycoses. 2022;65:65–70. doi: 10.1111/myc.13383. - DOI - PMC - PubMed
-
- Patel A., Agarwal R., Rudramurthy S.M., Shevkani M., Xess I., Sharma R., Savio J., Sethuraman N., Madan S., Shastri P., et al. Multicenter Epidemiologic Study of Coronavirus Disease–Associated Mucormycosis, India. Emerg. Infect. Dis. 2021;27:2349–2359. doi: 10.3201/eid2709.210934. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

