Resistance mechanisms and molecular epidemiology of Pseudomonas aeruginosa strains from patients with bronchiectasis
- PMID: 35323912
- PMCID: PMC9155640
- DOI: 10.1093/jac/dkac084
Resistance mechanisms and molecular epidemiology of Pseudomonas aeruginosa strains from patients with bronchiectasis
Abstract
Background: Non-cystic fibrosis bronchiectasis (BE) is a chronic structural lung condition that facilitates chronic colonization by different microorganisms and courses with recurrent respiratory infections and frequent exacerbations. One of the main pathogens involved in BE is Pseudomonas aeruginosa.
Objectives: To determine the molecular mechanisms of resistance and the molecular epidemiology of P. aeruginosa strains isolated from patients with BE.
Methods: A total of 43 strains of P. aeruginosa were isolated from the sputum of BE patients. Susceptibility to the following antimicrobials was analysed: ciprofloxacin, meropenem, imipenem, amikacin, tobramycin, aztreonam, piperacillin/tazobactam, ceftazidime, ceftazidime/avibactam, ceftolozane/tazobactam, cefepime and colistin. The resistance mechanisms present in each strain were assessed by PCR, sequencing and quantitative RT-PCR. Molecular epidemiology was determined by MLST. Phylogenetic analysis was carried out using the eBURST algorithm.
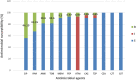
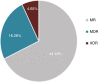
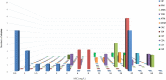
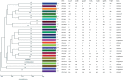
Results: High levels of resistance to ciprofloxacin (44.19%) were found. Mutations in the gyrA, gyrB, parC and parE genes were detected in ciprofloxacin-resistant P. aeruginosa strains. The number of mutated QRDR genes was related to increased MIC. Different β-lactamases were detected: blaOXA50, blaGES-2, blaIMI-2 and blaGIM-1. The aac(3)-Ia, aac(3)-Ic, aac(6″)-Ib and ant(2″)-Ia genes were associated with aminoglycoside-resistant strains. The gene expression analysis showed overproduction of the MexAB-OprM efflux system (46.5%) over the other efflux system. The most frequently detected clones were ST619, ST676, ST532 and ST109.
Conclusions: Resistance to first-line antimicrobials recommended in BE guidelines could threaten the treatment of BE and the eradication of P. aeruginosa, contributing to chronic infection.
© The Author(s) 2022. Published by Oxford University Press on behalf of British Society for Antimicrobial Chemotherapy.
Figures


Similar articles
-
Genomics and Susceptibility Profiles of Extensively Drug-Resistant Pseudomonas aeruginosa Isolates from Spain.Antimicrob Agents Chemother. 2017 Oct 24;61(11):e01589-17. doi: 10.1128/AAC.01589-17. Print 2017 Nov. Antimicrob Agents Chemother. 2017. PMID: 28874376 Free PMC article.
-
[Molecular epidemiology of beta-lactamases in ceftazidime-resistant Pseudomonas aeruginosa isolates].Mikrobiyol Bul. 2015 Apr;49(2):156-65. doi: 10.5578/mb.8901. Mikrobiyol Bul. 2015. PMID: 26167816 Turkish.
-
High incidence of MDR and XDR Pseudomonas aeruginosa isolates obtained from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial.J Antimicrob Chemother. 2019 May 1;74(5):1244-1252. doi: 10.1093/jac/dkz030. J Antimicrob Chemother. 2019. PMID: 30753505
-
Loss of activity of ceftazidime-avibactam due to MexAB-OprM efflux and overproduction of AmpC cephalosporinase in Pseudomonas aeruginosa isolated from patients suffering from cystic fibrosis.Int J Antimicrob Agents. 2018 Nov;52(5):697-701. doi: 10.1016/j.ijantimicag.2018.07.027. Epub 2018 Aug 3. Int J Antimicrob Agents. 2018. PMID: 30081137
-
The treatment of respiratory pseudomonas infection in cystic fibrosis: what drug and which way?Drugs. 2000 Nov;60(5):1053-64. doi: 10.2165/00003495-200060050-00006. Drugs. 2000. PMID: 11129122 Review.
Cited by
-
Quorum-sensing regulation of phenazine production heightens Pseudomonas aeruginosa resistance to ciprofloxacin.Antimicrob Agents Chemother. 2024 May 2;68(5):e0011824. doi: 10.1128/aac.00118-24. Epub 2024 Mar 25. Antimicrob Agents Chemother. 2024. PMID: 38526048 Free PMC article.
-
In Silico Molecular Analysis of Carbapenemase-Negative Carbapenem-Resistant Pseudomonas aeruginosa Strains in Greece.Microorganisms. 2024 Apr 16;12(4):805. doi: 10.3390/microorganisms12040805. Microorganisms. 2024. PMID: 38674749 Free PMC article.
-
Antibiotic Resistance and Virulence Determinants of Pseudomonas aeruginosa Isolates Cultured from Hydrocarbon-Contaminated Environmental Samples.Microorganisms. 2025 Mar 19;13(3):688. doi: 10.3390/microorganisms13030688. Microorganisms. 2025. PMID: 40142580 Free PMC article.
-
Virulence factors of Pseudomonas aeruginosa and immune response during exacerbations and stable phase in bronchiectasis.Sci Rep. 2025 Feb 22;15(1):6520. doi: 10.1038/s41598-025-91368-3. Sci Rep. 2025. PMID: 39987197 Free PMC article.
-
Rapid, sensitive, and user-friendly detection of Pseudomonas aeruginosa using the RPA/CRISPR/Cas12a system.BMC Infect Dis. 2024 Apr 30;24(1):458. doi: 10.1186/s12879-024-09348-3. BMC Infect Dis. 2024. PMID: 38689239 Free PMC article.
References
-
- Fernández-Barat L, Alcaraz-Serrano V, Amaro Ret al. . Pseudomonas aeruginosa in bronchiectasis. Semin Respir Crit Care Med 2021; 42: 587–94. - PubMed
-
- McDonnell MJ, Jary HR, Perry Aet al. . Non cystic fibrosis bronchiectasis: a longitudinal retrospective observational cohort study of Pseudomonas persistence and resistance. Respir Med 2015; 109: 716–26. - PubMed
-
- Del Barrio-Tofinõ E, Zamorano L, Cortes-Lara Set al. . Spanish nationwide survey on Pseudomonas aeruginosa antimicrobial resistance mechanisms and epidemiology. J Antimicrob Chemother 2019; 74: 1825–35. - PubMed
-
- Solé M, Fàbrega A, Cobos-Trigueros Net al. . In vivo evolution of resistance of Pseudomonas aeruginosa strains isolated from patients admitted to an intensive care unit: mechanisms of resistance and antimicrobial exposure. J Antimicrob Chemother 2015; 7: 3004–13. - PubMed
-
- Treepong P, Kos VN, Guyeux Cet al. . Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin Microbiol Infect 2018; 24: 258–66. - PubMed

