Reverse genetics systems for SARS-CoV-2
- PMID: 35324008
- PMCID: PMC9088479
- DOI: 10.1002/jmv.27738
Reverse genetics systems for SARS-CoV-2
Abstract
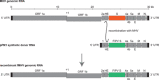
The ongoing pandemic of coronavirus disease 2019 (COVID-19) has caused severe public health crises and heavy economic losses. Limited knowledge about this deadly virus impairs our capacity to set up a toolkit against it. Thus, more studies on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) biology are urgently needed. Reverse genetics systems, including viral infectious clones and replicons, are powerful platforms for viral research projects, spanning many aspects such as the rescues of wild-type or mutant viral particles, the investigation of viral replication mechanism, the characterization of viral protein functions, and the studies on viral pathogenesis and antiviral drug development. The operations on viral infectious clones are strictly limited in the Biosafety Level 3 (BSL3) facilities, which are insufficient, especially during the pandemic. In contrast, the operation on the noninfectious replicon can be performed in Biosafety Level 2 (BSL2) facilities, which are widely available. After the outbreak of COVID-19, many reverse genetics systems for SARS-CoV-2, including infectious clones and replicons are developed and given plenty of options for researchers to pick up according to the requirement of their research works. In this review, we summarize the available reverse genetics systems for SARS-CoV-2, by highlighting the features of these systems, and provide a quick guide for researchers, especially those without ample experience in operating viral reverse genetics systems.
Keywords: BAC; CPER; SARS-CoV-2; TAR; reverse genetics systems.
© 2022 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Construction of a Noninfectious SARS-CoV-2 Replicon for Antiviral-Drug Testing and Gene Function Studies.J Virol. 2021 Aug 25;95(18):e0068721. doi: 10.1128/JVI.00687-21. Epub 2021 Aug 25. J Virol. 2021. PMID: 34191580 Free PMC article.
-
Reverse genetic systems of SARS-CoV-2 for antiviral research.Antiviral Res. 2023 Feb;210:105486. doi: 10.1016/j.antiviral.2022.105486. Epub 2022 Dec 22. Antiviral Res. 2023. PMID: 36657881 Free PMC article. Review.
-
Rescue of SARS-CoV-2 from a Single Bacterial Artificial Chromosome.mBio. 2020 Sep 25;11(5):e02168-20. doi: 10.1128/mBio.02168-20. mBio. 2020. PMID: 32978313 Free PMC article.
-
Versatile SARS-CoV-2 Reverse-Genetics Systems for the Study of Antiviral Resistance and Replication.Viruses. 2022 Jan 18;14(2):172. doi: 10.3390/v14020172. Viruses. 2022. PMID: 35215765 Free PMC article.
-
Reverse genetics systems for SARS-CoV-2: Development and applications.Virol Sin. 2023 Dec;38(6):837-850. doi: 10.1016/j.virs.2023.10.001. Epub 2023 Oct 11. Virol Sin. 2023. PMID: 37832720 Free PMC article. Review.
Cited by
-
Transformation-associated recombination (TAR) cloning and its applications for gene function; genome architecture and evolution; biotechnology and biomedicine.Oncotarget. 2023 Dec 22;14:1009-1033. doi: 10.18632/oncotarget.28546. Oncotarget. 2023. PMID: 38147065 Free PMC article. Review.
-
Highs and Lows in Calicivirus Reverse Genetics.Viruses. 2024 May 28;16(6):866. doi: 10.3390/v16060866. Viruses. 2024. PMID: 38932159 Free PMC article. Review.
-
Genome-Wide Analysis of the Indispensable Role of Non-structural Proteins in the Replication of SARS-CoV-2.Front Microbiol. 2022 Jun 1;13:907422. doi: 10.3389/fmicb.2022.907422. eCollection 2022. Front Microbiol. 2022. PMID: 35722274 Free PMC article.
-
Rapid assembly of SARS-CoV-2 genomes reveals attenuation of the Omicron BA.1 variant through NSP6.Nat Commun. 2023 Apr 21;14(1):2308. doi: 10.1038/s41467-023-37787-0. Nat Commun. 2023. PMID: 37085489 Free PMC article.
-
An optimized high-throughput SARS-CoV-2 dual reporter trans-complementation system for antiviral screening in vitro and in vivo.Virol Sin. 2024 Jun;39(3):447-458. doi: 10.1016/j.virs.2024.03.009. Epub 2024 Mar 26. Virol Sin. 2024. PMID: 38548102 Free PMC article.
References
-
- Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239‐1242. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

